Myosin: A Superfamily of Motor Proteins
| ✅ Paper Type: Free Essay | ✅ Subject: Biology |
| ✅ Wordcount: 3985 words | ✅ Published: 23 Sep 2019 |
Myosin: A Superfamily of Motor Proteins
Introduction
Every movement, from moving one’s fingers to looking around, is powered by myosin; myosin is one of only three proteins which are responsible for converting chemical energy into kinetic and mechanical work.1. It consists of a large family of motor proteins which are renowned for their various roles in muscle contraction. Along with that, the myosin family has an extensive range of functions in motility of eukaryotes. Overall, myosin uses chemical energy for motion. Myosin captures a molecule of ATP and breaks it, using the energy from ATP to perform the “power stroke” which will be further discussed along with the structure and mechanism. Myosin is the most important protein for muscle contraction and without it, our body’s biomechanics will fail to execute movement. At the same time, deficient functionality of myosin may resuIt in both biomechanical issues and cellular complications.
Myosin was first discovered in 1859, but 200 years ago its entire mechanical function was unknown. 80 years later, it was discovered that myosin consistently works with another protein, actin. The main discoveries were that myosin and actin consist of two different thin and thick filaments and that a muscle cell contraction takes place by a “sliding-like” motion of these filaments past one another. Today, the function of myosin can be further understood due to recent technology discoveries such as in-vitro assays and X-ray which; these methods introduce new insights into the way in which generic myosin motors convert chemical energy of ATP into the myosin’s mechanical work.2
In this paper, the importance of myosin contractile and cell division will be discussed. Also, there will be a focus on the protein superfamily components, key evolutionary residues, and the myosin-actin interactions with surrounding elements such as ATP and Pi. Lastly, the power stroke mechanism will be broken down to small details including the organic chemistry of ATP catalysis.
Biological Function
Together with actin, myosin is responsible for many different types of cell movements. Actin is a protein that alongside myosin forms contractile in muscle cell filaments and is involved in motion within other cells; structures and mechanisms of actin are less understood and therefore will not be further discussed in detail.3
Myosin is an example of a molecular motor, which is a protein that utilizes chemical energy in the form of ATP into mechanical energy which produces movement. The relevant movement to humans is muscle contraction. Besides muscle contraction, myosin plays a role in a variety of movements of non-muscle cells; such activities include cell division. In conclusion, myosin’s role in cell biology is vital.
First to be discussed are myosins responsible for the contraction of muscles. The unique specialty in the structure and function of muscle has made the muscle the main factor when studying movement at the molecular and cellular level. In vertebrates, there are three different types of muscle: skeletal muscle (responsible for all voluntary work), cardiac muscle (delivers blood from the heart) and smooth muscle (responsible for all involuntary movements of organs). Although they are not discussed in detail, it is essential to understand that the skeletal and cardiac muscles’ unique pattern in the cytoskeleton is what led to a profound examination of myosin.
Skeletal muscles are bundles of muscle fibers. Muscle fibers are large cells, often single, which are approximately 50 μm in diameter and usually a few centimeters long.4 Muscle fibers are formed by the fusion of groups of individual cells during cell development. Most of our cytoplasm is composed of myofibrils (cylindrical shaped bundles) of the myosin and actin filaments; myosin is a thick filament (~15 nm diameter), and actin is a thinner filament (~7 nm diameter). Each one of the myofibrils is uniquely organized as a long chain of contractile units, called sarcomere, which is responsible for the paralleled shape of the skeletal and cardiac muscle. To draw the connection, sarcomeres (~2.3 μm long) consist of several regions which are observable by electron microscopy, allowing the in-depth investigation into the mechanism of myosin.
To understand the idea of muscle contraction (which is driven by myosin), a unique contraction model must be explained: the sliding filament model. During muscle contraction, myosin filament slides past an actin filament so that the actin filament will move to another region (from region A to region H). Thus, muscle contraction occurs since myosin filament generates movement which is relative to the movement of actin. The basis to the movement in the molecular level is the interaction and binding of myosin with actin, thus allowing myosin to function as the single motor which drives the sliding of these filaments. The type of myosin that is present in the muscle is myosin II. Myosin II has two identical heavy chains which consists of two “heads”. Each one of the heads is composed of spherical region and a long tail with an α-helical shape. The α-helical tails twirl around each other in a coiled structure and form a dimer. Besides the two heavy chains, myosin II has two lighter chains which associate with the neck region (between the head and the tail) and complete the myosin II molecular structure.5 Besides binding to actin, myosin heads bind and hydrolyze ATP which generate the necessary energy for filament sliding. The conventional molecular mechanism suggests that filament sliding consists of a cycle which begins with ATP-absent myosin which is tightly bound to actin. Then, ATP binding disrupts the myosin attachment to actin and hydrolysis of ATP generates a conformational change in the myosin positioning. This change affects the neck region which is bound to the light chain and therefore displaces the myosin head by approximately 5 nm. ADP and Pi, which are the products of the hydrolysis of ATP are bound to the myosin head in the “pre-stroke” position. Then, the myosin head binds again at a new position on the actin and releases the ADP and Pi and essentially triggers the “power stroke,” in which the myosin head returns to its initial position, sliding the actin filament to another region of the sarcomere (chemical mechanism will be further discussed).6 Figure one illustrates the generic structure of myosin in both the pre-stroke position and its return the initial position, the rigor state. The power-stroke cycle mechanism will be further addressed, discussed, and illustrated.
The next group is a group of myosins responsible for contraction of non-muscle cells. Similar to muscle contraction, actin filaments possess a unique relationship with myosin II which generate the sliding-like motion which was previously discussed. The best example for a non-muscle cell myosin involvement is found in cytokinesis, which is the cell division into two mitosis. In animal cells, toward the end of mitosis, a “contractile ring” consisting of myosin II and actin is placed underneath the plasma membrane. The contraction of the ring pulls the plasma membrane inward, an action which narrows and pressures the center of the cell and pinching it in two. To clarify, following the nuclear division (mitosis), the contractile ring is the one pinching and dividing the cell in two. Figure two illustrates the cell division pinching pattern which takes place in cell division.
Besides myosin II, several other myosins are found in non-muscle cells. Opposed to myosin II, the other myosins (“unconventional”) are not involved in contraction. Some of them, however, can be related to different types of cell movements, such as transport of membrane vesicles and organelles (via the actin filaments). The most notoriously studied unconventional myosin is myosin I. Myosin I proteins, similarly to myosin II, contain a spherical head group which acts as a motor. The components of myosin I, however, are substantially smaller and lack the long tail of myosin organelles and structures such as membrane vesicles. An important function of myosin I include the formation of lateral arms that attach actin filaments to plasma membranes and move the plasma membrane toward the end of microvillus (actin-base bulge of the plasma membrane, common in absorption cells). Another role of myosin I is the transportation of vesicles and organelles via the actin filament and the movement of the plasma membrane during the uptake of large particles by a cell.
Other than Myosin I and II, there are 12 other classes of unconventional myosins (III through XIV).6 Some are similar to myosin II in their structure (two heads), and others are more similar to myosin I. Although most of the function of myosins III-XIV is undetermined, it is clear that myosins V and VI play a vital role in organelle movement and myosin III, VI, and VII play a role in sensory functions such as vision and hearing.
Structure, Mechanism, and Chemistry of Function
Myosins are a superfamily which binds to actin and serves as molecular motors which consists of at least thirteen myosin classes. The conserved residues of the motor domain are positioned in the framework which was provided by recent crystal structures of N-terminus (the free amine group at the end of a polypeptide); the usage of crystal structures helps to define the residues involved in binding the actin and ATP, hydrolysis, and conformational changes. Overall, there is a very poor conservation at the site which was thought to be involved in actin binding, but several other highly conserved residues have been identified.
The essential functionality residues in the myosin’s motor domain are almost exclusively found at the N-terminus, which is well conserved among all myosins. Besides the motor domain, a short, regulatory domain which is associated with the members of the calmodulin family (divalent cation binding proteins) was found in all myosins studied.7 The divalent cation binding proteins bind to the IQ-motif on myosin; the IQ-motif is a sequence of amino acids which usually follows a pattern of Ile–Gln–x–x–x–Arg–Gly–x–x–x–Arg. The term IQ is derived from the first two amino acids of the motif, namely isoleucine (I), and Glutamine (Q).7 What differs most myosins is the specificity at the C-terminal regions, which diverse in size and sequence; it is the C-terminal regions that determine the specific function of the myosin. The diversity of the C-terminal region indicates that the myosin family cannot be grouped and was therefore subdivided into three classes: conventional myosins (class II), single-headed unconventional myosins (class I), and the rest were divided into classes determined by order of discovery. Interestingly, as eukaryotic functionality evolved, so did the diversion in Myosins II sequences. Unlike prokaryotes, as multicellular organisms with special structures and tissues evolved, the functionality of myosins evolved as well (cardiac and skeletal myosins). Once skeletal, smooth, and cardiac muscle myosins II were introduced, the myosins II overall conserved its C-terminal residues and adopted a common backbone for similar actin-activated ATPase rates.8
The conservation of residues means that there is functional and structural importance. The key areas are the actin and nucleotide binding sites and the regions that involve conformational changes. The most conserved residues in myosin involve the binding of nucleotide, and most of the side chains are found in the well-conserved sequences GESGAGKT and NSSRFGK; these residues are illustrated in figure three. The conserved residues form the active site that fulfills a variety of roles in the power stroke which will be further discussed; the phosphate which binds to the head of the myosin is held firmly by interacting with glycine, alanine, and lysine. They make water-mediated contacts with the ATP phosphate which its ligands are responsible for the catalysis of the contractile cycle. Arginine is also relatively conserved and is associated with an “escape route” of a phosphate while ATP is hydrolyzed at the center of the motor domain.
Overall, the positions of the conserved residues which were examined in the motor domains provide with a hypothesis to their possible function. The residues related to ATP-binding and hydrolysis are now defined and are highly conserved. There is, however, very poor conservation in the proposed actin-binding site, implying that understanding the actin-myosin interface is much more complex than assumed.
To generate the power stroke, tension stimuli must occur in the nerve. This occurs by a stimulus which is spread down the alpha motor neuron. Then, acetylcholine (abbreviated as Ach) is released and crosses the synapse. The Ach opens up the Na+ and K+ channels to the sarcolemma. Then Na+ flows in the cell, while K+ flows out of the cell, creating the muscle fiber action potential. Once this occurs, Na+ goes down to the T-tubule system which causes Ca+ to be released from the sarcoplasmic reticulum.9 Ca++ then binds to troponin, and a change in the configuration of actin occurs which exposes its binding site, leading to the further discussion of the power stroke.10
The power stroke, or cross-bridge cycling, is the mechanism that generates contractile and is fully illustrated in figure four. Although there is a consensus on the general outline of the mechanism, much less is known on how it produces mechanical work. First, myosin binds initially through flexible loop regions near the catalytic site, followed by progressive and extensive hydrogen bonding. In the process, a piece of the protein is removed from the interaction with cell water during the bond formation. In cases in which protein binds similarly without conformational changes, it will cost 60-80kj/mol to form the bonds (twice than the observed number of myosin to actin); it suggests that the myosin head captures energy internally. When myosin moves into its tightly bound configuration, the energy deforms the myosin head, which applies force to the neck region. Most of this energy is channeled to mechanical work once the filaments slide past one another and myosin tightly bind to actin. Subsequently, Mg-ATP binds to myosin (a very high-affinity binding) and provides the necessary free-energy and thermodynamics to change the binding site to a lower affinity state, which allows further detachment. The binding of ATP substantially lowers the affinity of myosin for actin. Then, hydrolysis of ATP occurs which results in a subtle change in free energy and Pi and ADP are released from the ATPase site. The overall conversion of energy of the hydrolysis of ATP to the myosin-actin binding is about 50kj/mol and called chemomechanical transduction. The timing of ATP hydrolysis and ADP and Pi release in respect to the myosin-actin binding states and power stroke states is unknown; it is thought, however, that ATP hydrolysis often occurs after the transition of the actomyosin-ATP complex to its weaker-bound state. Then, the complex actomyosin-ATP, called A-M-ATP, remains weakly bound until it is detached by the filament movement. The released M-ADP-Pi (myosin, ADP, and Pi) has relatively moderate affinity for actin, and once reattached, it forms A-M-ADP-Pi and phosphate release occurs (Actomyosin, ADP, and a Pi which refers to a free phosphate); this generates a high-affinity state which is associated with the beginning of the famous power stroke. Once the structural change that occurred by the tight binding to actin produce high strain in the myosin head and forces a movement, ADP is released. Therefore, once power stroke is almost terminated, cross-bridges are typically in the actin-myosin state (rigor state) which is the tightest rigid state in which they remain until another ATP is available and re-alter the affinity. As long as [Ca2+] remains high, the cycle continues (if adequate ATP concertation and other internal conditions are met). The rate-limiting step is the release of Pi, and everything else, from the Pi release through ATP hydrolysis, happens quickly. Therefore, M-ADP-Pi and A-M-ADP-Pi state dominate. There is evidence that in high-speed contraction there might be many attachments and detachments per one hydrolysis. Therefore, it is hard to understand the cycle entirely.11
To fully understand the power stroke, the chemistry of ATP catalysis when binding to the myosin must be understood. Three reaction pathways, which are illustrated in figure five, were found to be associative with a pentavalent bipyramidal phosphorene transition state.12 A bond between Mg2+ cation coordination sphere and the switch-1 loop of the myosin (consists of Ser and Thr) must occur to break the product state; this step prepares the opening of the switch-1 loop (switch 1 loop is a central element which transmits information between the nucleotide binding pockets of myosin to future binding partners).13 The opening of the switch-1 loop is necessary for the release of hydrolysis products once power stroke occurs.
The first reaction path is the “Direct Path.” In the direct path, the activation of the attacking water happens by straight proton transfer of the water to the third phosphate of ATP. At transition state, the third phosphate adopts a geometry of trigonal bipyramidal. In the product state, the Mg shifts from the first and second phosphates and moves away from Ser237; therefore, breaking the bond with switch-1 loop. This action weakens the strong interaction that maintains the closure of switch-1 over the nucleotide. The second path is the “Ser236 Path.” Here, the Ser236 transfers the proton onto the third phosphate, and the “attacking water” changes orientation into an optimal position for proton transfer to Ser236. The third path is the “Ser181 Path.” Here, Ser181 transfers its proton to the third phosphate, and the “helper water” transfers a proton to the side chain of Ser181, and the “attacking water” moves its proton to the “helper water.” These three pathways which occur in similar likelihoods explain catalysis of ATPase and the following binding to the myosin head.
Conclusion
In this paper, both myosin II and other myosin classes were carefully examined. The paper introduced the purpose of myosin in biological aspects (cell division) and biomechanical aspects (the power stroke). Conserved key residues were introduced to examine what residual regions of the myosin have been conserved and what it is important. Through illustrations and detailed description, the structure of myosin, the organic chemical, and the biological components were introduced and clarified.
Overall, myosin II and its relation to the actin filament in contraction is fairly understood. Other myosins’ nuclear functions, however, are hardly understood. Dysfunctions of non-muscle myosins, such as mitosis myosin, have recently emerged as a critical regulator of genetically complex diseases, including various cancers, neuronal disorders, and vascular diseases. Nonetheless, the relationship and cause remain unknown.14 Broader focus and investigation on non-muscle myosins and other unconventional myosins must occur to decipher complex genetic diseases and potentially save human lives. With further research, the way we perceive non-muscle and contractile myosins may change forever. More profound understanding of conventional and unconventional myosin groups might not just prevent things from happening, but will provide with an opportunity to improve cell-division and contractile activity. Now, imagine a world in which muscle contractile activity is personally optimized to one’s needs and non-muscle myosins are understood and regulated to enhance control of cancerous mutations and neuronal and vascular diseases. Perhaps, this is what the future holds if myosin is better understood.
References
- Rayment, I. & Holden, H. M. The three-dimensional structure of a molecular motor. Trends Biochem. Sci. 19, 129–134 (1994).
- Stull, J. T. Myosin minireview series. J. Biol. Chem. 271, 15849 (1996).
- Dominguez, R. & Holmes, K. C. Actin structure and function. Annu. Rev. Biophys. 40, 169–86 (2011).
- Sweeney, H. L. & Houdusse, A. The motor mechanism of myosin V: insights for muscle contraction. doi:10.1098/rstb.2004.1576
- Cooper, G. M. The cell : a molecular approach. (ASM Press, 2000).
- Cooper, G. M. Actin, Myosin, and Cell Movement. (2000).
- Cope, M. J. T., Whisstock, J., Rayment, I. & Kendrick-Jones, J. Conservation within the myosin motor domain: implications for structure and function. Structure 4, 969–987 (1996).
- Uyeda, T. Q. P., Ruppel, K. M. & Spudich, J. A. Enzymatic activities correlate with chimaeric substitutions at the actin-binding face of myosin. Nature 368, 567–569 (1994).
- ROBERT L. POST, CSABA HEGYVAHY, A. S. I. Activation by Adenosine Triphosphate in the Phosphorylation Kinetics of Sodium and Potassium Ion Transport Adenosine Triphosphatase”. JOUHNAL OB R~OLOGICAL Chem. 247, 6530–6540 (1972).
- HUXLEY, A. F. Muscle structure and theories of contraction. Prog. Biophys. Biophys. Chem. 7, 255–318 (1957).
- N.V. BHAGAVAN. Muscle and Nonmuscle Contractile Systems. Medical Biochemistry (Fourth Edition) (2002). Available at: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/myosin-head. (Accessed: 28th November 2018)
- Sonja M. Schwarzl, ‡,§, Jeremy C. Smith, § and & Stefan Fischer*, ‡. Insights into the Chemomechanical Coupling of the Myosin Motor from Simulation of Its ATP Hydrolysis Mechanism†. (2006). doi:10.1021/BI052433Q
- Kintses, B. et al. Reversible movement of switch 1 loop of myosin determines actin interaction. EMBO J. 26, 265–74 (2007).
- Newell-Litwa, K. A., Horwitz, R. & Lamers, M. L. Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. (2015). doi:10.1242/dmm.022103
Figures

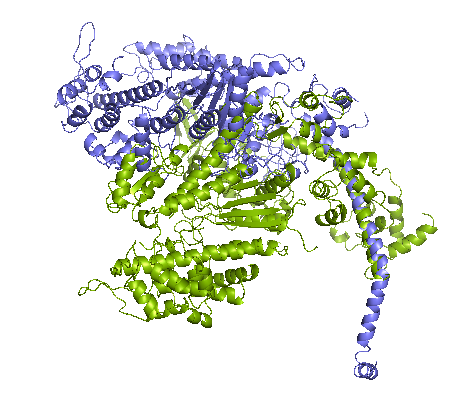
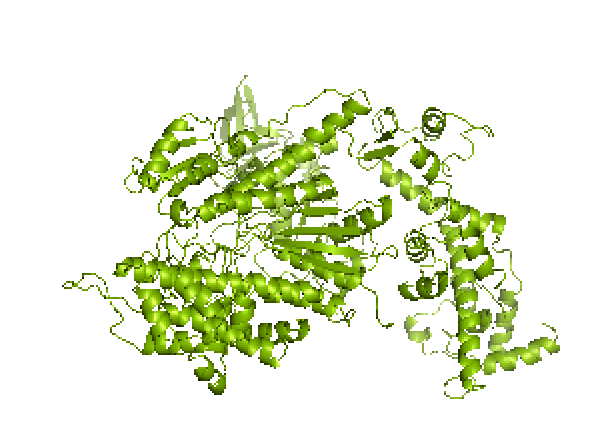
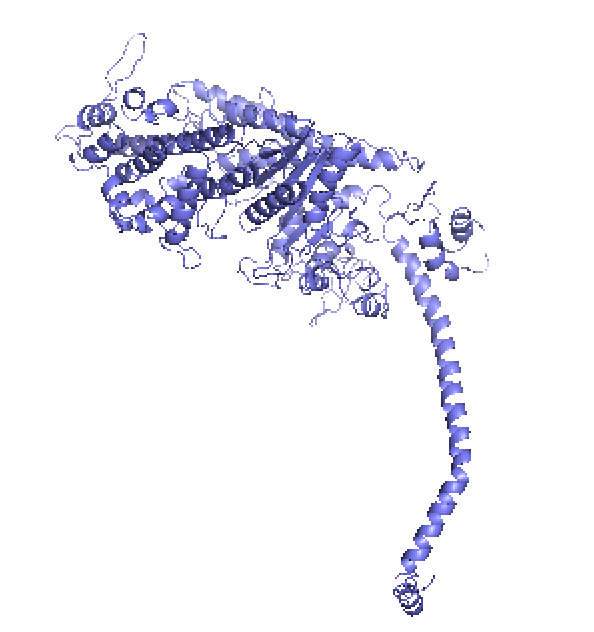
Figure 1. left: Structural overlays of the two states of the power stroke of myosin, pre-power stroke state and rigor state. The structure of the pre-stroke state is shown in green (after ATP cleaves), and the structure of the rigor state is shown in purple (after grabbing the actin filament and the release of phosphate). Right: the pre-stroke state (after ATP cleaves). Bottom: the rigor state (after grabbing the actin filament and the release of Pi).




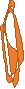
















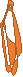
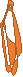
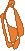
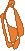
Figure 2. After mitosis, the contractile ring which consists of myosin II and actin divides the cell in two.


chicken 61 TVKTEGGE–TLTVKE—–DQVFSMNPPKYDKIEDMAMMTHLHEPAVLYNLKERYAAW
MYOSIN-VI 30 TIEPLNQKGKTFLALI—–NQVFPAEEDSKKDVEDNCSLMYLNEATLLHNIKVRYSKD
myosin-X 31 VFRTDYGQ–VFTYKQSTITHQKVTAMHPTNEEGVDDMASLTELHGGSIMYNLFQRYKRN
STRIATED 55 TVDIEGQE–SRQVKK—–DLLQQVNPPKYEKCEDMSNLTYLNDASVLHNLKQRYYAN
chicken 173 QSILITGESGAGKTVNTKRVIQYFATIAASGEKK-KEEQSGKMQGTLEDQIISANPLLEA
MYOSIN-VI 145 QSIIVSGESGAGKTENTKFVLRYLTESYGTG————-QDIDDRIVEANPLLEA
myosin-X 149 QCILISGESGAGKTESTKLILKFLSVISQQS—-LELSLKEKTSCVERAILESSPIMEA
STRIATED 167 QSMLITGESGAGKTENTKKVIAYYANVGAATPKPGKEAPTKEKKATLEDQVVQTNPVLEA
chicken 232 FGNAKTVRNDNSSRFGKFIRIHFGATGKLASADIETYLLEKSRVTFQLPAERSYHIFYQI
MYOSIN-VI 192 FGNAKTVRNNNSSRFGKFVEIHFNEKSSVVGGFVSHYLLEKSRICVQGKEERNYHIFYRL
myosin-X 205 FGNAKTVYNNNSSRFGKFVQLNICQKGNIQGGRIVDYLLEKNRVVRQNPGERNYHIFYAL
STRIATED 227 FGNAKTVRNDNSSRFGKFIRIHFGPMGKLAGADIETYLLEKARVISQQTLERSYHIFYQL
chicken 666 STHPHFVRCIIPNETKTPGAMEHELVLHQLRCNGVLEGIRICRKGFPSRVLYADFKQRYR
MYOSIN-VI 660 STGASFIRCIKPNLKMTSHHFEGAQILSQLQCSGMVSVLDLMQGGFPSRASFHELYNMYK
myosin-X 626 SSNPFFVRCIKPNMQKMPDQFDQAVVLNQLRYSGMLETVRIRKAGYAVRRPFQDFYKRYK
STRIATED 666 STQPHFVRCIIPNELKQPGVIDSGLVMHQLTCNGVLEGIRICRKGFPNRMVYPDFKQRYT
Figure 3. Multiple sequence alignment of myosin proteins. Sequences for chicken myosin chicken II, green fluorescent myosin-VI (myosin-VI), human myosin-X (myosin-X), and human muscle striated myosin (striated) are shown in multiple sequence alignment. Residues shown in black are universally conserved. Residues shown in grey/brown represent conservative mutations.


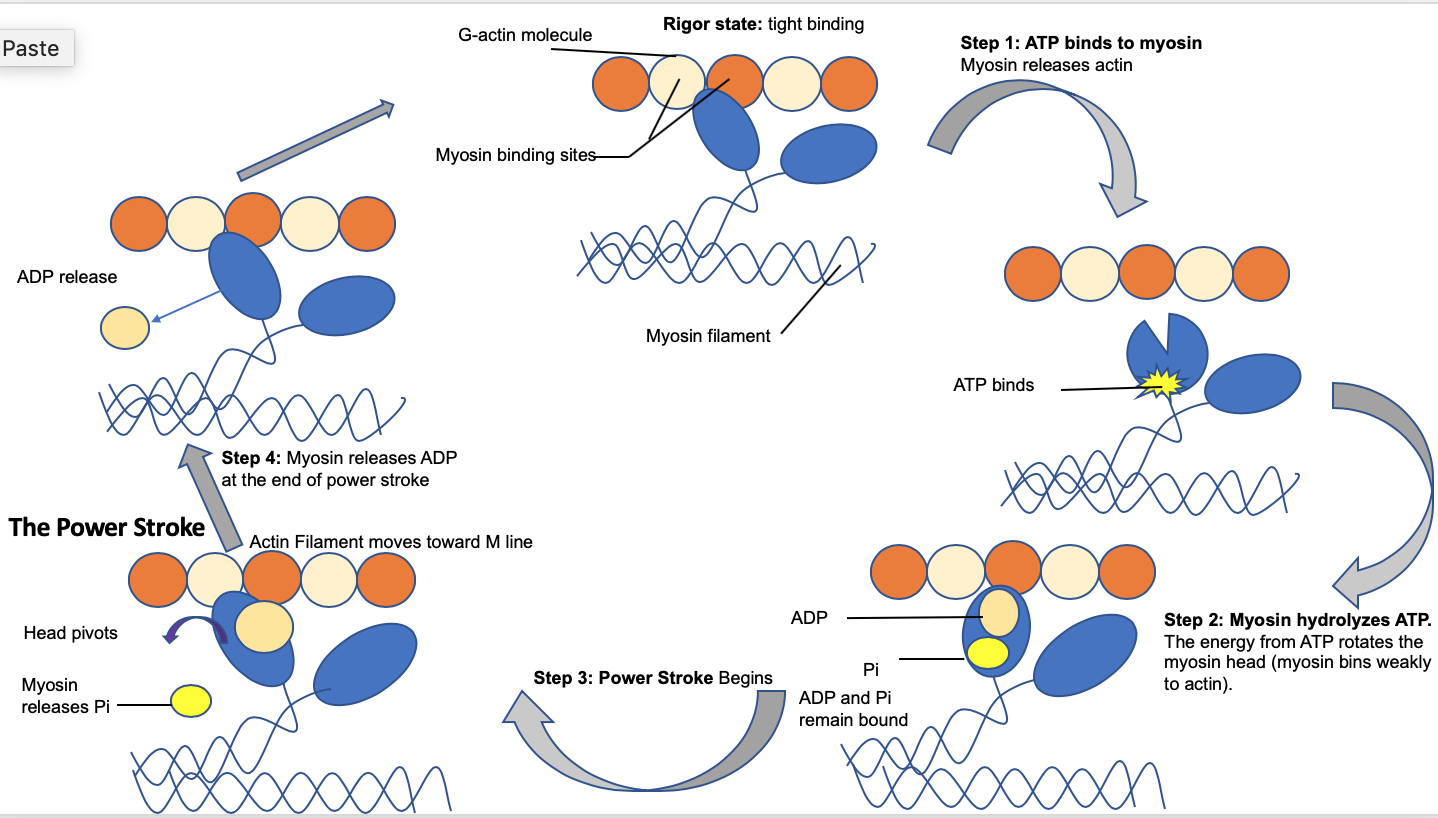
Figure 4. The power stroke mechanism is driven by cleavage of ATP and phosphate which cause the shift between the pre-power stroke state, the bent/flexed form attached to actin, and the rigor state.
Step 1: Starting a new cycle, the myosin head lacks a bound ATP, and it is connected to the actin filament in an unstable conformation, called “rigor conformation.” Step 2: ATP binds to the myosin head domain and leads to a conformational shift in the binding site to actin which reduces the affinity for actin and causes myosin head to release the actin filament completely. Then, ATP binds causes a massive conformational shift in the “lever arm” or “neck” of the myosin that bends the myosin head. Then, ATP is hydrolyzed, leaving a Pi and ADP connected to myosin. Step 3: The myosin head, attached to ADP and Pi, makes slight contact with the actin filament and a minute conformational change occurs to the myosin neck which leads to the release of Pi Step 4: The release of Pi leads to a tighter binding between the myosin and actin filament and triggers the power stroke (the blueprint of force-generating step). Then, actin forces myosin to return to its original conformation and release ADP. Step 5: once myosin reverts to its starting position, ADP is fully released, and the myosin head remains bound to the actin filament, generating the next cycle.


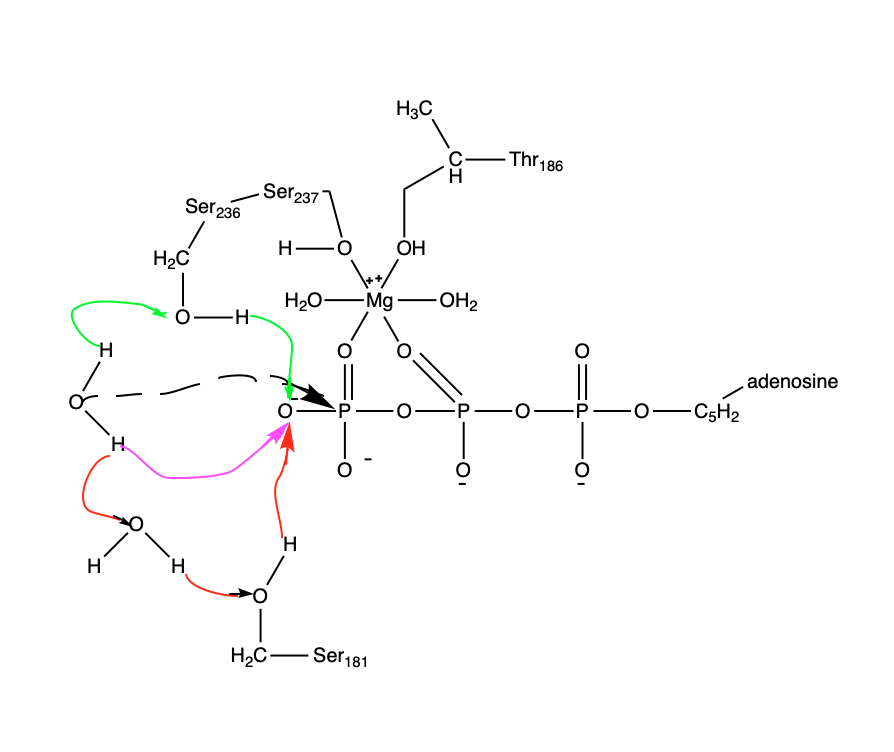
Figure 5: Potential reaction pathways for ATP hydrolysis. Three different mechanisms of the water activation are demonstrated: pink arrow corresponds to “direct path,” green arrows to “Ser236 path” and red arrows to the
“Ser181 path”. The three pathways of water activation are found to happen equally. The dashed arrow demonstrates an attack of the “activated water”’ on the third-phosphate of ATP (gamma-phosphate). The top water molecule is the “attacking water” and the bottom one is the “helper water.”

Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allDMCA / Removal Request
If you are the original writer of this essay and no longer wish to have your work published on UKEssays.com then please click the following link to email our support team:
Request essay removal


