Applications of Metagenomics to Medicine
| ✅ Paper Type: Free Essay | ✅ Subject: Sciences |
| ✅ Wordcount: 3284 words | ✅ Published: 23 Sep 2019 |
Introduction to Metagenomics
What is Metagenomics?
Genomics is the interdisciplinary field of science focusing on structure, function, evolution, mapping, and editing of genes. Historically, microbial genomics involved the isolation of microbes for genetic analysis through culturing methods to identify the genetic profile of species within a given sample. In recent years, Metagenomics has risen as a new frontier both as a research field and approach to circumvent the unculturability and genomic diversity of most microbes, the biggest roadblocks to advances in clinical and environmental microbiology.[1] The discipline of metagenomics represents the study of population genomics at the level of microorganisms, referencing the notion that a collection of genes from a given environment can be analyzed in a way analogous to that of a single genome, offering a powerful lens into the microbial world that has the potential to revolutionize the clinical sciences.[2]
History of Metagenomics
 Conventional genomics began with the culturing of identifiable cells as a DNA source for analysis. However, early researchers investigating the preliminary notions of metagenomics hypothesized that the major weakness of conventional genomics is that groups of microorganisms cannot be analyzed if they cannot be cultured, and thus cannot be sequenced for genomic analysis. To overcome this hurdle in the field of genomics, the bacteria-specific 16S rRNA sequence was the main focus of early metagenomics as this sequence was relatively short, often conserved within a species, and generally different between species.[3] This led to the first report of isolating and cloning of bulk DNA from an environmental sample, published by Pace and colleagues in 1991.[4]
Conventional genomics began with the culturing of identifiable cells as a DNA source for analysis. However, early researchers investigating the preliminary notions of metagenomics hypothesized that the major weakness of conventional genomics is that groups of microorganisms cannot be analyzed if they cannot be cultured, and thus cannot be sequenced for genomic analysis. To overcome this hurdle in the field of genomics, the bacteria-specific 16S rRNA sequence was the main focus of early metagenomics as this sequence was relatively short, often conserved within a species, and generally different between species.[3] This led to the first report of isolating and cloning of bulk DNA from an environmental sample, published by Pace and colleagues in 1991.[4]
The Metagenomic Analysis Strategy
Bioinformatics

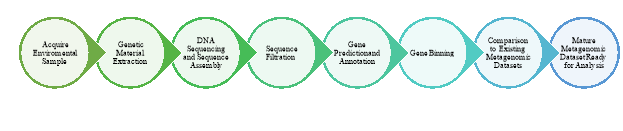 Figure 1: Metagenomic Analysis Workflow
Figure 1: Metagenomic Analysis Workflow
DNA Sequencing
One of the strongest potentials of metagenomics when compared to conventional genomics lies is the ability to detect trends and correlations indicative of interactions between the microbial world and the environment. In present day, metagenomic analyses are affordable and accessible to the average microbiology project, allowing for the generation of massive sequence outputs.[5] The first step a metagenomic analysis after acquiring of a sample involves the sequencing of DNA. Currently, two main approaches are utilized to generate the bulk metagenomic data: Shotgun metagenomics and High-throughput sequencing. Shotgun metagenomics involves the random sheering of DNA following its extraction, resulting in the formation of many short sequences which are then reconstructed into a consensus sequence. Shotgun metagenomics provides information about which organisms are present and what metabolic processes are possible in the community.[6] High-throughput sequencing allows for the sequencing of multiple DNA molecules in parallel, enabling hundreds of millions of DNA molecules to be sequenced at a time. This advantage allows HTS to be used to create large data sets, generating more comprehensive insights into the cellular genomic and transcriptomic signatures of various diseases and developmental stages.[7] These sequencing techniques used in metagenomics bypasses the cloning and culturing requirements of traditional genomic studies before sequencing can be conducted, removing one of the main biases and bottlenecks in microbial environmental sampling.
Consequently, the data that is generated by metagenomic experiments are both enormous and inherently noisy, containing fragmented data representing as many as 10,000 species of microbes.[8] Bioinformatics is used to acquire relevant biological information from the generated following sequencing so that the metagenomic dataset generated at the end of the experiment can be analysed. Contaminating eukaryotic genomic DNA sequences and other non-relevant genomic data to the study are removed, allowing for the assembly of DNA sequences identifying microbes, and their relevant abundance, genes, and gene functions. Coding regions of the genes of interest in assembled contigs are annotated based on homology or by known intrinsic features of sequences from suspected related organisms. Once gene annotation has occurred, genomic binning is conducted to measure species diversity within the produced metagenomic dataset.[9] Genomic binning can be conducted in two ways. Similarity-based binning is used to rapidly search for phylogenetic markers or otherwise similar sequences in existing public databases, and composition-based binning using intrinsic features of the sequence.[8] Once binning has occurred, the metagenomic dataset is compared with existing or known datasets through the use of programs such as MEGAN (MEta Genomic ANalyzer) to explore taxonomic diversification of the dataset, comparing the resulting sequences with gene sequences from GenBank in NCBI.[40] Gene annotations provide the “what”, while measurements of species diversity provide the “who”.[10] Metagenomic datasets derived from a given community (e.g. patients sharing with similar urologic diseases), can identify microbial groups which are responsible for conferring specific characteristics of given environment, and is the main goal of metagenomic studies in the clinical sciences, providing additional insight into the function of complex microbial communities and their role in host health.[11]
Metagenomics in Medicine
Clinical Relevance of Metagenomics
Currently diagnosis of a vast majority of microbial diseases are carried out using traditional culture-based methods. In a clinical context, culture-based methods can fail to isolate disease-causing organism[12] and are time consuming and labor intensive.[13] Although metagenomics has not reached to the point of being a standard practice, utilizing a metagenomic approach clinically has the potential to identify and characterize bacterial and viral pathogens,[14] and data generated can be utilized as functional information to understand complex infections at a genetic level as well as how members of the microbiome contribute to disease through interactions with host physiology.[14]
Three themes have emerged from the application of microbial ecology to clinical microbiology, where metagenomics can have its applications. Within the clinical laboratory, most bacteria cannot be isolated in pure culture, so molecular methods will report a wider range of organisms than culture and are generally more sensitive.[15] Differences in host-associated microbial communities can influence the balance between health and disease in conditions not normally thought of as microbial or infectious in origin (e.g. inflammatory bowel disease, cancer or obesity).[16] Lastly, developments in infection research have suggested that interactions between organisms in a community can influence disease outcomes and in some cases it might even be appropriate to treat a whole microbial community as a pathogenic entity, as opposed to the notion that a single pathogen causes a single disease.[18]
The Potential of Clinical Metagenomics Application
Prior to a discussion on how metagenomics can be used for diagnostic purposes in the clinical setting, it is worth reviewing the problems associated with existing diagnostic approaches. Microbial diagnostic techniques currently used in the clinical setting today were developed over a century ago (the detection and characterization of bacteria through microscopy and gram staining, and culturing of clinical isolates).[19-20] In situations where microscopy is cumbersome, unrewarding or difficulties in culturing is encountered, culture-independent approaches to pathogen detection are used, including immunoassays and detection of nucleic acid sequences.[21] Although practical, these approaches are generally target-specific and thus lack the ability to detect unsuspected pathogens, resulting a battery of tests that may have to be applied to each sample, each of which requires optimization and standardization.[20]
Clinical diagnostic metagenomics brings the promise of an open-ended, assumption-free one-size-fits-all workflow that could be applied to any specimen to detect any kind of pathogen.[20] Given the rapid development of tools targeted for pathogen identification, and likely future improvements in the ease, throughput and cost-effectiveness of sequencing, twinned with commoditization of laboratory and informatics workflows, one can foresee a tipping point when a unified automated metagenomics-based workflow will start to compete with the plethora of methods currently in use in the diagnostic laboratory, while also delivering additional useful information (e.g. genomic epidemiology, antimicrobial resistance, virulence). Given the rapid development of tools targeted for pathogen identification, it is feasible that metagenomics will play a key role in the clinical laboratory in the near future.
A Future for Clinical Diagnostic Metagenomics
However, one must ask, what stands in the way of the application of metagenomics clinically? Diagnostic metagenomics is still currently in its infancy. A study in 2013 using metagenomics to investigate diarrhea samples that were positive for Shiga-Toxigenic E. coli showed a sensitivity of only 67% compared to culture.[14] One of foremost problems with diagnostic metagenomics is the handling, interpreting, and making use of the tremendous amounts of data afforded by metagenomics.[22] Metagenomics to be used in the clinical setting not only requires the knowledge of the Manual of Clinical Microbiology but also all of environmental microbiology, International Journal of Systematic and Evolutionary Microbiology and the entirety of the NCBI taxonomy database.[22] This will require the formation of a not currently existent workforce in which health organizations will need to budget and develop when they already currently have functional and profit-generating microbiology teams using traditional methods for diagnosis. Another major challenge is the cost of a single diagnosis. One of the main benefits of the current clinical microbiology testing paradigm is that it is cost-effective.[22] For comparison, current clinical microbiological markups in the clinical microbiology lab convert <$5 of reagents and minutes of technician time into $200–500 reimbursements, compared to current metagenomic testing prices in the low four figures ($2000–3000) per sample.[22] Hence, for diagnostic metagenomic to be implemented widespread, the costs associated with such testing will need to be reduced.
However, when the implementation of clinical diagnostic metagenomics does occur, and as metagenomics becomes cheaper and faster, it will become possible to serially characterize human microbiomes to investigate for disease associations. This could lead to personalized medicine for diseases that related to the host genome and resident microbiomes, and to personalized treatments such as the use of narrow-spectrum antibiotics to reduce disruption of the microbiome or specific probiotics to restore an individual’s microbiome to a healthy state.[23] Although the widespread implementation of diagnostic metagenomics has not occurred yet, this has not stopped researchers from applying such techniques at a research level. Diagnostic metagenomics has been used to diagnose C.jejuni from fecal samples[12], E.coli from urine samples[24], and lymphocytic choriomeningitis virus in fatal infections in transplant recipients.[25] These studies shows the potential of clinical diagnostic metagenomics in disease diagnosis.
Metagenomics in Urology
Microbiome of the Urogenital Tract
The metagenomic relevance in the discipline of urologic medicine is due to the emerging evidence in the microbiota’s role in maintaining urinary health. Studies of the urinary microbiota have identified remarkable differences between healthy populations and those with urologic diseases.[26] Microorganisms at sites distal to the kidney, bladder and urethra are likely to have a profound effect on urologic health, both positive and negative, owing to their metabolic output and other contributions.[26] Connections between the gut microbiota and renal stone formation have already been discovered.[26]
However, the relationship between these actively metabolizing organisms and urogenital health has yet to be completely elucidated. Given the role of the kidneys and bladder in filtration and storage of waste, respectively, microbial profiles and microbial metabolites of the gut and other organs might influence the urinary microbiota, and alterations might affect urinary homeostasis. Whether the microbiomes of these sites are predictive of the risk of urological disease or malfunction is unclear at the moment.[26] Due to the importance of the urinary microbiota to both an individual’s health and disease manifestation, metagenomic research of the urinary and gut microbiomes is warranted and can lead to insights on how microbiomes influence host health and urologic diseases.
A Role of Metagenomics in Urology
Conventional microbiological methods are inadequate to fully determine the diversity of bacteria that are present in urine.[27] To understand how metagenomics can be applied to urology, we must first investigate the current urological tests clinically available. Urinary tract infections are among the most common bacterial infections[28], and are usually classified as uncomplicated and complicated, but more recently also by risk factors and severity grading depending on the clinical presentation[29]. The diagnosis requires clinical symptoms and evidence of living bacteria in the urine, usually quantified by numbers of colony forming units per milliliter (CFU/ml). Culture tests and urine microscopy have been the gold standard for diagnosing UTI.[29] However, no fixed bacterial count has been considered conclusive for significant bacteriuria in all kinds of UTIs and under all circumstances[31]. The currently used urine culture method for diagnosis is limited to detecting easily culturable aerobic bacteria only and not fastidious and anaerobic bacteria.[32] The underlying idea has always been that urine from healthy subjects is sterile and a negative or positive urine culture has usually been taken as discriminative for an infection to be absent or present, respectively.[32]
Metagenomic approaches appears to more comprehensively and quantitatively describe the urinary microbiome. [34] Recent metagenomic research how shown a broad range of even non-culturable bacteria can be detected in the” sterile” bladder urine in healthy individuals.[32] Thus, sterile urine may be a myth and recent metagenomic findings on the urine microbiome encourage a discussion to redefine the criteria for urinary tract infections and non-infectious urological disorders with similar symptoms.[32] Diagnostic metagenomics used clinically in the future may enable clinicians to detect a wide range of fastidious and anaerobic and even non-culturable bacteria in the “sterile” bladder urine of healthy individuals as well as in patients with different urological disorders.[32]
Metagenomic analysis may prove to be a key factor in determining the key microbiome community in preventing urolithiasis. Urolithiasis (Kidney Stones) affect up to 10% of the Canadian population and can lead to pain, hospitalization, lost of time at work, and surgery. Compositional analysis of kidney stones has revealed that over 80% of stones consist of calcium and oxalate derived from dietary and bodily processes. Individuals who have high levels of oxalate in their urine have a greater tendency to generate stones. Some patients, despite reducing their oxalate intake, still have high amounts in the urine[35-38]. The gut microbiome has been referred to as a metabolic organ that communicates with, and complements, our own human metabolic apparatus, and has been associated with associated with diabetes, obesity, cardiovascular disease and urologic diseases.[34] Evidence has shown that urolithiasis patients may have distinct gut microbial profiles when compared to control subjects.[34] A diagnostic metagenomic application could be a plausible method of approach in analysing patient microbiomes to determine whether the presence or absence of gut microbiome members may result in urolithiasis. Further treatment plans can be envisioned from this, drawing inspirations from gut microbiome modulations, (e.g. fecal transplantation). With previous studies in the transfer of gut microbiome from diabetic-free patients into diabetic patients and improvements in insulin sensitivity as a result[34], gut microbiome manipulation may represent a novel preventative treatment for urolithiasis patients in a similar fashion. In the future, systematic, prospective use of metagenomic tools for disease diagnosis in urologic medicine may shed light on the role of unknown and unconventional microorganisms in the urinary tract or gut that may have clinical relevance towards urologic disease manifestation.
References
- National Research Council (US) Committee on Metagenomics: Challenges and Functional Applications. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet. Washington (DC): National Academies Press (US); 2007.
- Marchesi JR. Metagenomics: current innovations and future trends. Future Microbiol 2012;7:813–814.
- Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69(2):330-9.
- Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173(14):4371-8.
- Bertelli C, Greub G. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin Microbiol Infect. 2013;19:803-13.
- Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Computational meta’omics for microbial community studies. Mol Syst Biol. 2013;9:666.
- Churko JM, Mantalas GL, Snyder MP, Wu JC. Overview of high throughput sequencing technologies to elucidate molecular pathways in cardiovascular diseases. Circ Res. 2013;112(12):1613-23.
- Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6(2):e1000667.
- Thomas T, Gilbert J, Meyer F. Metagenomics – a guide from sampling to data analysis. Microb Inform Exp. 2012;2(1):3.
- Konopka, A. What is microbial community ecology?. The ISME Journal. 2009;3(11):1223–30.
- Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14(4):169-81.
- Nakamura S, Maeda N, Miron IM, et al. Metagenomic diagnosis of bacterial infections. Emerg Infect Dis. 2008;14(11):1784-6.
- Pallen MJ, Loman NJ, Penn CW. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol. 2010;13(5):625-31.
- Loman NJ, Constantinidou C, Christner M, et al. A Culture-Independent Sequence-Based Metagenomics Approach to the Investigation of an Outbreak of Shiga-Toxigenic Escherichia coli O104:H4. JAMA. 2013;309(14):1502–10.
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annual Reviews in Microbiology. 2003;57, 369-94.
- Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3(3):14.
- Rogers GB, Stressmann FA, Walker AW, et al. Lung infections in cystic fibrosis: deriving clinical insight from microbial complexity. Expert Reviews in Molecular Diagnostics. 2010;10(2):187–96.
- Wang B, Yao M, Lv L, et al. The human microbiota in health and disease. Engineering. 2017;3:71–82.
- Gram, HC. Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten. Fortschritte der Medizin 2, 1884;185–9.
- Pallen MJ. Diagnostic metagenomics: potential applications to bacterial, viral and parasitic infections. Parasitology. 2014;141(14):1856-62.
- Morré SA, Sillekens P, Jacobs MV, et al. RNA amplification by nucleic acid sequence-based amplification with an internal standard enables reliable detection of Chlamydia trachomatis in cervical scrapings and urine samples. J Clin Microbiol. 1996;34(12):3108-14.
- Greninger AL. The challenge of diagnostic metagenomics. Expert review of molecular diagnostics. 2018;18(7):605-15.
- Miller RR, Montoya V, Gardy JL, Patrick DM, Tang P. Metagenomics for pathogen detection in public health. Genome Med. 2013;5(9):81.
- Hasman H, Saputra D, Sicheritz-Ponten T, et al. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol. 2014;52(1):139-46.
- Palacios G, Druce J, Du L, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. New England Journal of Medicine 2008;358(10):991-8.
- Whiteside SA, Razvi H, Dave S, et al. The microbiome of the urinary tract: a role beyond infection. Nat Rev Urol. 2015;12(2):81–90.
- Lewis DA, Brown R, Williams J, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol. 2013;3:41.
- Colgan R; Williams M. Diagnosis and treatment of acute uncomplicated cystitis. American Family Physician. 2011;84(7):771–6.
- Johansen TE, Botto H, Cek M et al. Critical review of current definitions of urinary tract infections and proposal of an EAU/ESIU classification system. Int J Antimicrob Agents. 2011;38 Suppl:64-70.
- Grabe M., Bartoletti R., Bjerklund Johansen T.E., et al. EAU Guidelines on urological infections. European association of urology. 2015.
- Hooton TM, Roberts PL, Cox ME, Stapleton AE. Voided midstream urine culture and acute cystitis in premenopausal women. N Engl J Med. 2013;369(20):1883-91.
- Smelov V, Naber K, Bjerklund Johansen TE. Letter to the Editor: Diagnostic Criteria in Urological Diseases do not Always Match with Findings by Extended Culture Techniques and Metagenomic Sequencing of 16S rDNA. Open Microbiol J. 2016;10:23-6.
- Moustafa A, Li W, Singh H, et al. Microbial metagenome of urinary tract infection. Sci Rep. 2018;8(1):4333.
- Stern JM, Moazami S, Qiu Y et al. Evidence for a distinct gut microbiome in kidney stone formers compared to non-stone formers. Urolithiasis. 2016;44(5):399-407.
- Heilberg IP, Schor N. Renal stone disease: Causes, evaluation and medical treatment. Arq Bras Endocrinol Metabol. 2006;50(4):823-31.
- Sakhaee K. Nephrolithiasis as a systemic disorder. Curr Opin Nephrol Hypertens. 2008;17(3):304-9.
- Sakhaee K. Recent advances in the pathophysiology of nephrolithiasis. Kidney Int. 2009;75(6):585-95.
- Worcester EM, Coe FL. Nephrolithiasis. Prim Care. 2008;35(2):369-91.
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377-86.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allDMCA / Removal Request
If you are the original writer of this essay and no longer wish to have your work published on UKEssays.com then please click the following link to email our support team:
Request essay removal


