Functions and Applications of miRNA and miRNA Sponge Technology
Info: 14557 words (58 pages) Dissertation
Published: 24th Feb 2022
Abstract
The advent of next generation sequencing technology has allowed for the vast world of ncRNA to be explored. Previously thought to have redundant function within the cell, modern insights have revealed this class of RNA to be key in posttranscriptional regulation. This is especially true of miRNA and miRNA sponge which are relatively recent discoveries. The function of miRNA is to silence gene expression through a variety of mechanisms and protein interactions. On the other hand, miRNA sponge act as regulators of miRNA. They bind miRNA and act as decoys which prevents miRNA from binding to their target. Advances in sequencing technology has not only increased understanding of the biogenesis of these RNA sequences, but has also increased our ability to utilise them. The gene silencing abilities of miRNA have been examined for numerous applications including use as therapeutics and as a means of cell engineering. Indeed, inhibition of miRNA themselves is becoming increasingly studied as we understand how miRNA expression affects cell development and contributes to disease. This is where miRNA sponge have a major role to play. This review outlines current understanding regarding the generation, natural functions and applications of this class of RNA.
Contents
List of Abbrieviations
1. Introduction
2. Types of ncRNA involved in RNAi
2.1 MicroRNA (miRNA)
2.2 Small interfering RNA (siRNA)
2.3 piwi interacting RNA (piRNA)
2.4 Micro RNA Sponge (miRNA sponge)
3. Biosynthesis of MicroRNA (miRNA)
3.1 Transcription by poll II
3.2 The Microprocessor Complex
3.3 Exportin 5
3.4 Cleavage by Dicer
3.5 Argonaute and the RISC complex
3.6. miRNA targeting and silencing of mRNA
4. Natural Functions and Applications of miRNA
4.1 Beta Cell Regulators:
4.2 Role in Multiple Sclerosis
4.3 miRNA mimics:
4.4 miRNA in Biotechnology:
5. MicroRNA Sponge Biogenesis
5.1. Linear miRNA Sponge:
5.2 Circular miRNA Sponge Biogenesis
6. Natural Functions and Applications of miRNA Sponge
6.1. Roles in disease:
6.2 Possible Therapeutics:
6.3 Role in CHO Cell Engineering:
7. Conclusion
Experimental Design
Bibliography
List of Abbrieviations:
| ncRNA: non-coding RNA
RNAi: RNA interference miRNA: microRNA miRNA sponge: microRNA sponge siRNA: small interfering RNA piRNA: piwi interacting RNA pol II: RNA Polymerase II pri-miRNA: primary microRNA Dgcr8: DiGeorge Critical Region 8 dsRNA: double stranded RNA SBE: SMAD binding elements lncRNA: long non-coding RNA RBP: RNA Binding Proteins 3’ UTR: 3’ Untranslated Region RBP1: Ran Binding Protein 1 TRBP: Trans-activation Response Binding Protein DTE: difficult to express miR-NT: negative control miRNA |
circRNA: Circular RNA
RISC: RNA-induced silencing complex MBS: multiple binding sites RFP: Red Fluorescent Protein CHO: Chinese Hamster Ovary isomiRNA: isoform miRNA dsRBP: double stranded RNA binding protein AGO2: Argonaute 2 ssRNA: single stranded RNA PABP: poly A binding protein 375KO: miR-375 gene knocked out TG375: transgenic mice with enhanced miR-375 expression STZ: streptozotocin MS: Multiple Sclerosis SIRT1: silent information regulator 1 PTEN: Phosphatase and Tensin Homolog |
1. Introduction
Next generation sequencing techniques such as Illumina sequencing has identified many types of non-coding RNA (ncRNA). As these RNA do not code for protein expression, the role of this class of RNA within the cell is only in recent times becoming understood. The primary function of this subclass is the inhibition of gene expression through their reactions with a plethora of proteins. These proteins can halt mRNA translation or trigger transcript degradation in a process known as RNA interference (RNAi). There are many different types of ncRNA and while some classes actively inhibit gene expression, others act to inhibit other types of ncRNA (Jeck et al. 2013).
Two types of ncRNA which are attracting increasing levels of focus are microRNA (miRNA) and microRNA sponge (miRNA sponge). Knowledge of miRNA and miRNA sponge is still very much in its infancy with the first reports of miRNAs only occurring in the 1990s. This was when Ambros and colleagues detailed the gene silencing abilities of the short RNA sequences generated by lin-4 in C. Elegans (Lee, Feinbaum and Ambros 1993). The miRNA sponge are an even more recent discovery with their initial documentation in Arabidopsis thaliana. In this plant, the mRNA transcript of IPS1 was a target of miR-399 but was not cleaved by the miRNA. (Franco-Zorrilla et al. 2007).
Since their initial discoveries, an understanding of the biogenesis as well as the many applications of these ncRNA has grown. This review aims to look at different types of ncRNA with focus on the generation and function of miRNA and miRNA sponge. The biogenesis of these ncRNA, in particular, has opened-up new understanding of gene regulation within the cell. This has led to studies in their expression patterns in disease with either overexpression or low expression of a miRNA or miRNA sponge often a significant factor in causing disease (Li, Meng and Yang 2017, Liu et al. 2017). Such observations have raised the possibility of using miRNA or miRNA sponge as therapeutics (Jung et al. 2015, Chorn et al. 2012). In addition, this review also looks at how they proving to be potent tools in the bioprocessing industry for efficient generation of protein products (Fischer et al. 2014, Sanchez et al. 2014).
2. Types of ncRNA involved in RNAi
2.1 MicroRNA (miRNA)
MicroRNA are short RNA sequences involved in gene silencing through their interaction with RISC (Chi, Hannon and Darnell 2012). They were first discovered in the early 1990s however it wasn’t until the work of Pasquinelli et al. (2000) that their potential for inhibiting gene expression across species was first witnessed. Since then, miRNA have been identified as critical players in gene regulation and have been subject to intense study. Their applications range from biomarkers (Latreille et al. 2015) to therapeutics (Matsui, Prakash and Corey 2016) to CHO cell engineering (Fischer et al. 2014).
2.2 Small interfering RNA (siRNA)
Small interfering RNA are very similar to miRNA in the sense that they also silence gene expression. However, siRNA differ from miRNA in the fact they target a specific gene rather than a variety of genes like miRNA (Lam et al. 2015). In addition to this, siRNA are not encoded for in the genome. They are generally derived from exogenous sources such as viruses or transgenes. Although, they still react with the same machinery as miRNA as they are processed by both Drosha and Dicer (Carthew and Sontheimer 2009). Several siRNA based drugs are in clinical trials which target lung disease, cancer and inflammation and have been reviewed by Chakraborty et al. (2017).
2.3 piwi interacting RNA (piRNA)
Piwi interacting RNA are a subclass of interfering RNA which are only expressed in animal reproductive organs. They are 24 to 31 nucleotides in length and they differ from other small RNAs as they do not require processing by the Dicer enzyme (Xiol et al. 2012). They target transposons which are mobile elements of DNA. To do this, they bind with cleavage proteins such as PIWI, Aubergine and Argonaute-3. PIWI is involved in germline stability whilst Aubergine is required to generate healthy oocytes (Brennecke et al. 2007).
The piRNA are initially transcribed from a cluster as one long transcript before undergoing primary processing. These transcripts contain cis-elements which guide the transcripts to their processing machinery. Any sequence downstream of these elements are cleaved to form primary piRNA with a preference for a uridine at their 5’ end. These piRNA bind with their PIWI and Aubergine proteins before targeting transposons for cleavage. Secondary piRNA are then formed from cleavage of a target transcript. This is where one of the cleavage proteins with primary piRNA bound cleaves a target into two sequences and the second sequence, downstream of the cleavage site, enters piRNA biogenesis. Its 5’end is formed by PIWI cleavage. This secondary fragment then binds to Argonaute-3 which cleaves primary piRNA transcripts (Yang et al. 2016). This is referred to as the “ping-pong” model where the secondary piRNA and associated cleavage protein cleave the initial transcript of a primary pi-RNA to form more mature piRNAs (Zhang et al. 2011).
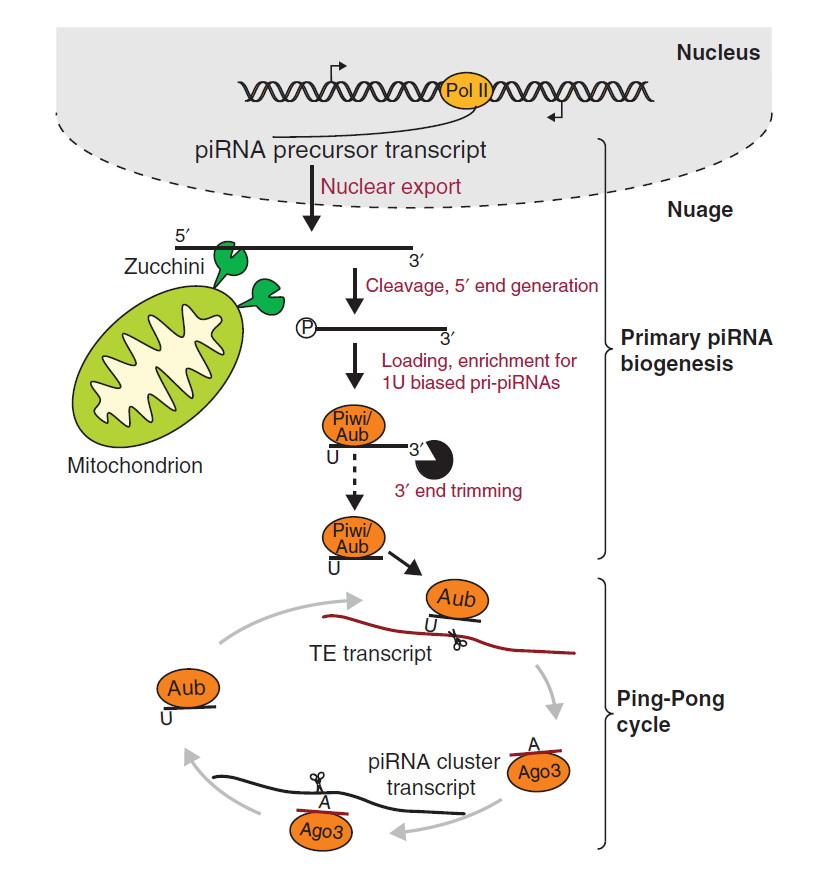
Fig 1: Primary piRNA precursors are transcribed in the nucleus before being exported from the nucleus. Their 5’ ends then undergo cleavage by Zucchini which is present on mitochondrial membranes. This produces intermediate piRNA with 5’ ends having uracil as the first base. After 3’ end trimming, they are loaded into Piwi or Aubergine which go onto cleave transposons (TE transcript). This produces secondary piRNA which load into Argonaute-3 (Ago3) and they in turn target primary piRNA precursors for cleavage. This creates a piRNA biogenesis loop known as the “ping-pong” model (adapted from Le Thomas, Toth and Aravin 2014).
2.4 Micro RNA Sponge (miRNA sponge)
Micro RNA Sponge are RNA sequences which can compete with natural targets for miRNA binding. They can exist in both linear and circular form and be encoded for by the genome either naturally or through transgene expression (Sanchez et al. 2014, Hansen et al. 2013). They are not to be confused with anti-miRNA oligonucleotides which are synthetic antisense RNA sequences that bind to miRNA in order to inhibit their target binding. While this is a heavily studied method of miRNA inhibition, the use of anti-miRNA has its limitations. The antisense RNA oligonucleotides are prone to degradation by endonucleases and exonucleases. In addition, they may cause toxicity to the cell (Lennox et al. 2013). This is where focus has been given to designing miRNA sponge as they can be stably incorporated into and expressed by the cell’s genome. They can have multiple binding sites (MBS) for either a single miRNA or multiple miRNA with high specificity for their targets. Also, their expression can also be inducible (Jung et al. 2015).
3. Biosynthesis of MicroRNA (miRNA)
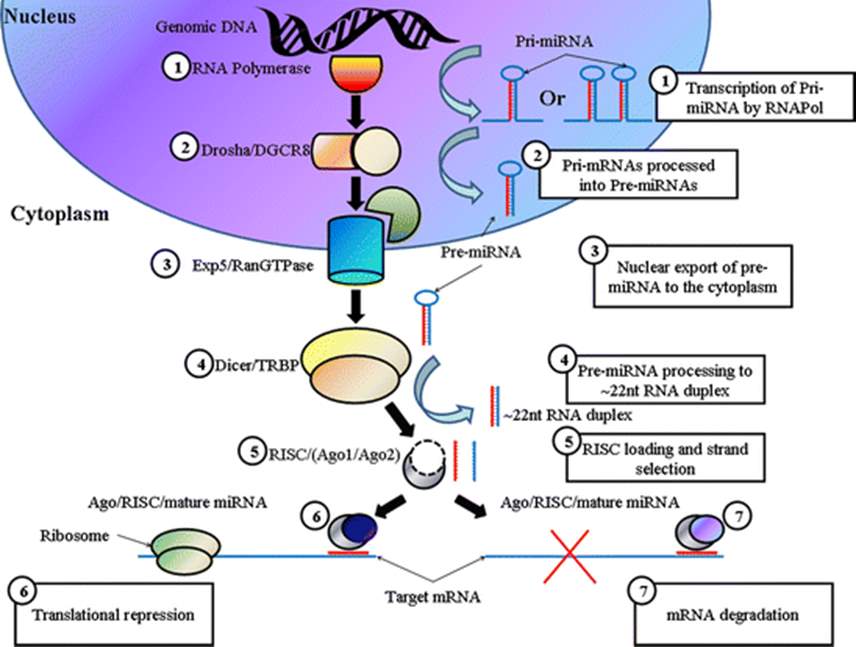
Fig 2: MircoRNA is initially transcribed in the nucleus as pri-miRNA which is then cleaved to pre-miRNA by Drosha. This is exported from the nucleus where it is further cleaved by Dicer. The double stranded miRNA can then bind to Argonaute where it is unwound to form a single strand. This forms the RISC complex which in turn targets mRNA to induce gene silencing (Barron et al. 2011B).
3.1 Transcription by poll II
MicroRNAs are initially transcribed in the nucleus by RNA polymerase II (pol II) and the initial transcript is referred to as the primary microRNA (pri-miRNA) transcript (see Fig 2) (Blahna and Hata 2012). Like most RNA transcripts, they are subject to 5’end capping with a 7-methyl-guanosine cap and poly adenylation at the 3’ end (Cai, Hagedorn and Cullen 2004). Both these RNA processing features function to maintain stability of the nascent RNA transcript as well as aid in exporting the nascent RNA from the nucleus (Daffis et al. 2010, Batisse et al. 2009). The pri-miRNA is then cleaved by an enzyme known as Drosha which forms stem-loop precursors that are approximately 70 nucleotides in length (see Fig 2). These are referred to as pre-miRNAs (Yoontae Lee et al. 2003).
3.2 The Microprocessor Complex
Drosha is part of a large protein complex called the microprocessor complex. One of the proteins in this complex is an RNase III enzyme which cleaves the pri-miRNA into pre-miRNA (Wu et al. 2012). Another component is DiGeorge critical region 8 (Dgcr8). Dgcr8 contains two double stranded RNA (dsRNA) binding domains and it is necessary for enabling Drosha to target pri-miRNA ( Han et al. 2006, Gregory et al. 2004). The Dgcr8 gene is monoallelic and deletion of it results in DiGeorge Syndrome. This causes numerous developmental defects and can result in heart and liver failure (Chen et al. 2012).
Drosha and Dgcr8 also play significant roles in regulating each other’s activity. This is so as to prevent excessive miRNA production (Han et al. 2006). The Drosha-Dgcr8 complex cleaves Dcgr8 mRNA transcripts preventing excess Dcgr8 expression. In addition, Drosha protein requires Dcgr8 to stabilise it. Low concentrations of Drosha result in a lack of microprocessor activity. This in turn stimulates Dgcr8 expression which upregulates Drosha (Han et al. 2009).
Other important elements of the microprocessor complex proteins are the Dead Box Helicase proteins: p68 and p72. These helicases are central to the unwinding of the pri-miRNA to make it accessible for cleavage by Drosha (Fukuda et al. 2007). Other functions include their ability to recruit SMAD proteins which are essential to the further maturation of a subset of miRNA. The stem-loop of the pri-miRNA for this class of miRNA contain conserved sequences called SMAD binding elements (SBE). This allows the microprocessor complex to bind to these pri-miRNA. The SMAD proteins then act as signal transducers for Transforming Growth Factor β and Bone Morphogenic Protein which enable microprocessor cleavage (Davis et al. 2010, Davis et al. 2008,). As well as SMAD proteins, binding of the tumour suppressor p53 to p68 allows for correct microprocessor assembly (Suzuki et al. 2009).
One of the main factors in guiding the microprocessor to the pri-miRNA is the long non-coding RNA (lncRNA) NEAT1. NEAT1 acts as a scaffold for RNA-binding proteins (RBP) such as the NONO-PSF dimer which can bind pri-miRNA. It also has a 3’ Untranslated Region (3’UTR) and many stem loops. These features are believed to be what attracts the microprocessor to it, and by extension, the microprocessor to the pri-miRNA (Jiang et al. 2017).
3.3 Exportin 5
The majority of pre-miRNAs are exported from the nucleus to the cytoplasm through the nuclear pore called Exportin 5 (see Fig 2). However, some miRNAs have been shown to use alternative nuclear pores such as the miR-320 family which are exported by Exportin 1. They have a cap that is specific to the family which is recognised by Exportin 1 through an adapter molecule called PHAX. It has also been demonstrated that knockout of Exportin 5 in cells does not totally halt miRNA biogenesis. This suggests that other export mechanisms may be involved (Kim, Kim and Kim 2016).
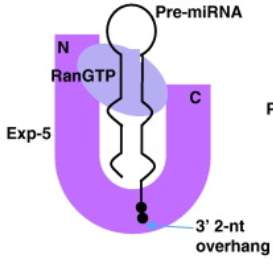
Fig 3: Exportin 5 forms a glove like structure around pre-miRNA and Ran GTP. HEAT repeats in the N-terminal and the C-terminal form the outer “thumbs” of the glove. Electrostatic interactions keep the stem loop of the pre-miRNA stable in the glove. HEAT repeats 6-19 of Exportin 5 are involved in binding to pre-miRNA. HEAT repeats 12-15 at the bottom of the glove connect and anchor the pre-miRNA to inner part of Exportin 5 (Okada et al. 2009). (Figure adapted from Lee et al. 2011)
For Exportin 5 to be able to export the pre-miRNA, it must be bound to a GTPase called RanGTP. In the absence of an export substrate (i.e. pre-miRNA), RanGTP has little affinity for Exportin 5. However, when pre-miRNA is present it readily binds to the Exportin 5 (see Fig 3). Ran Binding Protein 1 (RanBP1) then binds to the RanGTP-Exportin complex which allows a protein called RanGAP1 to access the complex. RanGAP1 induces hydrolysis of RanGTP and this causes the Exportin 5 complex to dissociate; thereby releasing pre-miRNA into the cytoplasm (Kim, Kim and Kim 2016, Maurer et al. 2001).
3.4 Cleavage by Dicer
Once in the cytoplasm, the pre-miRNAs are cleaved from their stem loop by an RNase III enzyme, called Dicer. This forms double stranded RNAs that are ~22 nucleotides in length (Saito et al. 2005). To do this, the open helical ends of the pre-miRNA bind to a RNA binding domain called PAZ (see Fig 4). PAZ recognizes and binds to the phosphorylated 5’ end of the pre-miRNA (Park et al. 2011). PAZ is separated from the RNase III catalytic site by the “platform domain” (see Fig 4). The RNase III catalytic site is composed of two RNase III domains in humans which dimerise (MacRae, Zhou and Doudna 2007).
When binding, the stem loop of the pre-miRNA is positioned adjacent to the Helicase domains (see Fig 4). The RNase III site in Dicer cleaves the dsRNA approximately 22 nucleotides from the open helical end as this is the distance between PAZ and the RNase III site (Lau et al. 2012).
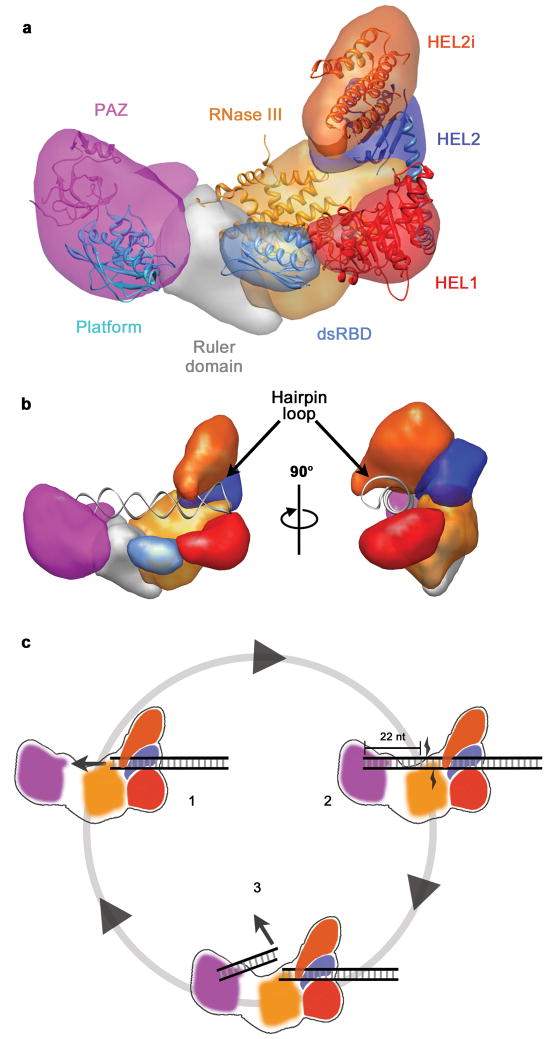
Fig 4: (a) The positional layout of the PAZ, RNase III and Helicase domains (HEL1, HEL2, HEL2i) in Dicer. (b) Stem loop held in position by Helicase domains adjacent to RNase III domain. (c) cleavage of pre-miRNA 22 nucleotides from open helical end (adapted from Lau et al. 2012).
Dicer interacts with a variety of proteins when processing pre-miRNAs and these vary from species to species. For example, in Drosophila it interacts with R2D2 and Loquacious (Lim et al. 2016). An ortholog of Loquacious in mammals is Trans-activation Response Binding Protein (TRBP). TRBP has been shown to alter the processing rates of pre-miRNA substrates depending on the structure of the pre-miRNA that Dicer acts on. In substrates with an unpaired nucleotide bulge at the cleavage site, TRBP inhibits processing. However, in substrates where all nucleotides at the cleavage site are paired, processing is enhanced. This is not the only feature which determines processing rate by the Dicer-TRBP complex. RNA loop size on the pre-miRNA also plays a role with larger RNA loops having been shown to have better processing rates than smaller loops. This attributed to Dicer having stronger binding affinity to larger loops. TRBP is also understood to have a role in creating isoform miRNA (isomiRNA) which are miRNA derived from the same pre-miRNA substrate that differ in length by one or two nucleotides (Lee and Doudna 2012, Chendrimada et al. 2005).
Another protein which binds to Dicer in mammals is protein activator of PKR (PACT). PACT binds to Dicer in the same manner as TRBP as it uses the C-Terminal dsRNA binding domain (dsRBD in Fig 4). PACT’s two N-terminal dsRNA binding domains are responsible for binding to dsRNA and recruiting it to Dicer. Whilst it has been observed to have a much less significant effect on the processing rate of pre-miRNA substrates compared to TRBP, PACT-binding does have a role in substrate selection. In particular, PACT causes Dicer to favour pre-miRNA processing over pre-siRNA processing (Lee et al. 2013).
However, not all miRNA are processed by Dicer for maturation. One such miRNA is miR-451. Once processed by Drosha, miR-451 is directly loaded into Argonaute which catalyses its cleavage (Cheloufi et al. 2010).
3.5 Argonaute and the RISC complex
Once cleaved, the dsRNA then leaves Dicer before re-associating with it (see Fig 5). Dicer is then bound to a double stranded RNA binding protein (dsRBP) such as TRBP which facilitates re-association of the dsRNA to Dicer in a different orientation. The dsRBP is located at the Helicase domains of Dicer and activates “asymmetry sensing”. This is where one end of the dsRNA has a 5’ end that is less thermodynamically stable than the other end. Dicer identifies the less thermodynamically stable end and positions the dsRNA such that the strand with this end is loaded as the guide strand into a protein called Argonaute 2 (AGO2) (Noland, Ma and Doudna 2011). The are 4 forms of mammalian Argonaute but AGO2 is the most common (Juvvuna et al. 2012).
AGO2 contains a PAZ domain which allows it to bind to the 3’ end of the dsRNA. PAZ binds to the 2-nucleotide 3’ end overhang of the dsRNA (see Fig 5). The MID domain of AGO2 binds the phosphorylated 5’ end of the dsRNA (Frank, Sonenberg and Nagar 2010). In this way, Dicer loads the dsRNA into Ago 2 (Gu et al. 2012, Ma, Ye and Patel 2004). However, AGO2 can’t accept the dsRNA in its usual conformation. It requires a chaperone called HSP90 (see Fig 5) which uses ATP hydrolysis to leave AGO2 open for insertion of the dsRNA (Martinez and Gregory 2013).
To form mature RISC, the passenger strand must be removed from the dsRNA (Park and Shin 2015).
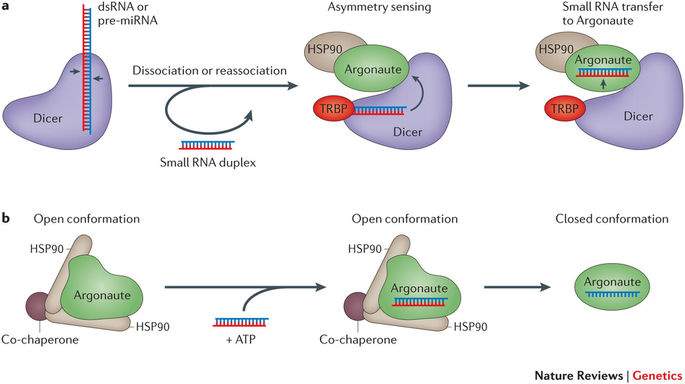
Fig 5: (a) dsRNA is removed from Dicer and re-inserts in a different region. TRBP activates Dicer so that it senses the asymmetry of the dsRNA and aligns so that the guide strand is loaded into Argonaute. (b) HSP90 hydrolises ATP to allow Argonaute to be loaded with the dsRNA. The passenger strand is then removed to form mature RISC (adapted from Meister 2013).
The method by which the passenger strand is removed from the dsRNA-RISC loading complex is not fully understood. However, there are two possible mechanisms for dsRNA unwinding. These are the slicer dependent pathway and slicer independent pathway. In the slicer dependent mechanism, AGO2 acts as a nuclease and unwinds the dsRNA. This pathway is unique to AGO2. However, this mechanism is used mostly for cleaving siRNA duplexes. For a miRNA duplex, the slicer independent pathway is followed. This is where the passenger strand is viewed as a target of the guide strand. The 3’ end of the guide strand is held in position by the PAZ domain of AGO2. Another domain of Argonaute protein, the N-domain, prevents base pairing beyond position 16 in the guide strand. These features, in combination with the mismatches in the miRNA duplex, are believed to enable AGO2 to undergo a conformational change that results in passenger strand dissociation (Park and Shin 2015, Gu et al. 2012, Kwak and Tomari 2012).
3.6. miRNA targeting and silencing of mRNA
The mature RISC with single stranded RNA (ssRNA) guide binds to mRNA with its “seed” region which is located 2-8 nucleotides from the 5’ end (Chi, Hannon and Darnell 2012). AGO2 is unique in the sense it can alter the 3’ end of miRNA by mediating the addition or removal of 1-2 nucleotides which forms isomiRNA. This is believed to change not only the binding affinity of the miRNA to some of its targets, but also which targets it preferentially binds (Juvvuna et al. 2012).
The “seed” sequence binds mostly to the 3’ UTR of the mRNA transcript however, it has been shown to bind to the 5’ UTR in instances where complementarity exists. Complementarity between miRNA seed sequences and the 3’ UTR does not have to be perfect for binding to occur. (Lytle, Yario and Steitz 2007).
Upon binding to their target sequence, miRNAs can suppress translation and trigger mRNA decay. The GW182 protein family has a prominent role in this; particularly their mammalian paralogs TNRC6A, TNRC6B and TNRC6C. This protein family have been shown to inhibit protein synthesis by inhibiting translation (Zipprich et al. 2009). GW182 is also central to the degradation of mRNA by deadenylation which is the removal of the poly-A tail. GW182 binds with CCR4-NOT deadenylase complex through its CNOT1 subunit. GW182 also has a PAM2 motif which binds PAN2-PAN3 deadenylase (Fabian et al. 2011).
In addition to this, the GW182 proteins trigger poly A binding protein (PABP) to dissociate from mRNA. This is despite evidence suggesting that PABP initially aid in the binding of RISC to mRNA (Flamand et al. 2016, Zekri, Kuzuoğlu-Öztürk and Izaurralde 2013). By removing the poly-A tail, the mRNA can be signalled for cleavage by endonuclease proteins. In this way, the mRNA is not translated and the gene expressing the mRNA transcript is silenced (Horman et al. 2013, Chen and Shyu 2011)
4. Natural Functions and Applications of miRNA
4.1 Beta Cell Regulators
There are many natural regulatory functions in the cell for miRNA. An example of one regulatory miRNA is miR-375. Whilst this miR-375 is found throughout the body, it is highly expressed within pancreatic Beta cells (Latreille et al. 2015). Studies have been carried out which determined the exact role that miR-375 plays within the cell and whether its expression level can be indicative of a disorder. Poy et al. (2009) and Latreille et. Al (2015) looked at whether expression levels of miR-375 are altered during type 1 and type 2 diabetes. Type 1 diabetes is where the body attacks its own beta cells and results in lower insulin production and hyperglycaemia (Mathis, Vence and Benoist 2001). Type 2 diabetes is where cells become insensitive to insulin and this causes beta cells to highly produce insulin to compensate (Nolan, Damm and Prentki 2011). It was postulated that these disease types may relate to miR-375’s functions in the cell. If so, miR-375 concentration could be a potential biomarker for either disease state.
Mice were generated with the miR-375 gene knocked out (375KO) as well as transgenic mice with enhanced miR-375 expression (TG375). The 375KO mice were observed to be hyperglycaemic as well as having decreased beta cell mass. However, pancreatic alpha cell mass had increased (Poy et al. 2009). The decrease in beta cell mass is attributed to miR-375 having a role in suppressing key transcripts which alter beta cell formation. These targets include Caveolin 1, Inhibitor of DNA binding 3 and Smarca2 which all have regulatory roles in the growth of beta cells. By allowing the uncontrolled proliferation of these miR-375 targets, beta cell formation did not occur in the correct manner (Poy et al. 2009).
Long term glucose injection revealed that these 375KO mice had lower levels of insulin production (than wild type mice) which resulted in the hyperglycaemia and diabetes. However, cross-breeding these mice with TG375 mice restored blood glucose levels to near wild type levels. These mice were referred to as beta-rescued mice. (Latreille et al. 2015).
Surprisingly, the TG375 mice were indistinguishable from wild type mice both in terms of beta cell mass and insulin secretion. This has been attributed to beta cells possibly already having high levels of control where miR-375 is already in excess (Latreille et al. 2015).
When mice models are treated with streptozotocin (STZ) (which induces beta cell death), serum from these mice display higher levels of miR-375. Beta cell injury is what occurs during Type 1 diabetes which means miR-375 could be used as a biomarker for Type 1 diabetes. Studies have since shown that miR-375 is elevated in Type 1 diabetes patients (mouse and human) and is a valid biomarker (see Table1).
Table 1: miR375 as a Biomarker (adapted from Latreille et al. 2015)
4.2 Role in Multiple Sclerosis
The role of miRNAs as a regulator means they themselves could be significant contributors to disease states. Multiple Sclerosis (MS) is a neurodegenerative disorder of the Central Nervous System where nerve cells become demyelinated. This often results in serious physical and possible mental disability for the sufferer (Liu et al. 2017).
While the exact factors which cause MS are not fully understood, Th17 cells, which are thought to be stimulators of both chronic and autoimmune inflammation, are believed to be one of the contributors to MS. The miRNA, miR-590, initiates the proliferation of CD4+ T cells to Th17 cells through its silencing of the Tob1 gene. This gene codes for proteins that prevent interleukin-17A expression. Interleukin-17A induces differentiation to the Th17 cell type which means Tob1 expression is necessary to prevent aberrant Th17 formation. In a study by Liu et al. (2017), patients with MS had their periphery blood cells as well as their cerebrospinal fluid examined for miR-590 concentrations. The presence of miR-590 in MS patients was significantly increased in these samples which confirms unregulated expression of miR-590 as a contributor to MS (Liu et al. 2017). It also demonstrates the key roles miRNA play, both in terms of preventing and causing, abnormal cell proliferation and function which underline many diseases.
4.3 miRNA mimics
With more and more observations of miRNA regulation of cell structure and function, synthetic miRNA or “miRNA mimics” have now become commercially available for in vitro and in vivo applications. RNA sequences can be created through chemical synthesis and this has led to the creation of sequences which are virtually identical to naturally occurring miRNA (Pradère, Halloy and Hall 2017). As they are chemically synthesised, manufacturers may also add modifications which enhance miRNA performance. These modifications include where the RNA sugar has a 2-O-Methyl substitution or has molecules like cholesterol linked to it to improve in vivo specificity (Krützfeldt et al. 2005). Other modifications can give augmented thermodynamic features or increased efficiency of RNA-induced silencing complex (RISC) incorporation. Once synthesised, the miRNA mimics can be delivered to the cell through a variety of vectors; such as liposomes (Goldgraben et al. 2016).
Conventionally, mimics have been double stranded when added to the cell. However, for miRNA mimics to realise their potential as therapeutics, there has been a significant push towards the generation of single stranded miRNA mimics. This is because double stranded RNAs are less able to enter cells in vivo and are susceptible to “off-target” effects due to the passenger strand (Matsui, Prakash and Corey 2016). This is where the passenger strand may inadvertently target and silence mRNA strands which were not the intended target (Jackson et al. 2003).
Many miRNA mimics have been created with potential therapeutic benefits. One particular mimic is the miR-34a mimic. This miRNA is a constituent of the p53 pathway in cells. The p53 protein is a transcription factor which regulates apoptosis through the expression of a cluster of miRNAs. One of these miRNAs is miR-34a which targets the silent information regulator 1 (SIRT1) mRNA transcript. SIRT1 dictates cell apoptosis by deacetylating certain molecular targets (including p53 itself) in response to stress (Yamakuchi, Ferlito and Lowenstein 2008). A disturbance to the p53 expression pathway can result in tumour formation as cells fail to undergo apoptosis after being damaged (Luo et al. 2001). Both double stranded and single stranded mimics of miR-34a have been synthesised and they have demonstrated that in the treatment of colon cancer, in vitro, tumour progression is halted when miR-34a silences SIRT1 expression (Matsui, Prakash and Corey 2016, Yamakuchi, Ferlito and Lowenstein 2008).
Mimics have also been generated for miR-124 and miR-122, in vitro, which have a role in suppressing liver cancer. They too have been modified to enhance their activity over their natural counterparts. These modifications included 5’ phosphorylation and 2’-fluoro substitution of the RNA oligonucleotides (Chorn et al. 2012).
As reviewed by Rupaimoole and Slack (2017), there are numerous miRNA mimic based therapies in clinical trials for diseases such as Hepatitis C virus and cancer. However, one must not disregard that the disadvantages of miRNA mimics are numerous. The chemical modifications introduced by manufacturers may mislead experiments involving mimics. In addition, the mechanism of action followed by a mimic may not be same as its natural counterpart i.e. the mimic may inhibit translation whilst the natural miRNA causes mRNA degradation. There has also been evidence that miRNA mimics can cause uncontrolled changes to gene expression within the cell. In terms of effect, as they are added to the cell exogenously using plasmids or lentivirus (or the previously mentioned liposomes), their action within the cell is only temporary (Jin et al. 2015).
4.4 miRNA in Biotechnology
The rise of miRNA technology has seen its applications tested in a variety of industrial settings. The production of biotherapeutics requires host cell lines (e.g. CHO cell lines) which can be genetically engineered to produce therapeutic proteins. As cell line development is an expensive and time-consuming process, miRNAs offer an avenue to greater cell line stability and enhanced production quality. For miRNAs to be expressed in CHO cell lines, they must be identified, sequenced and PCR amplified before recombination with a plasmid (Barron et al. 2011A). These plasmids are available commercially and can encode a variety of selection markers such as G418 resistance. The plasmids are transfected into the cells and the cells are grown in selective media containing agents such as G418. This screens for CHO cells which have stably incorporated the plasmid into their genome. The cells, which have previously been transfected to stably express a transgenic reporter protein such as a GFP, SEAP or an antibody, are then separated into individual wells and the best expressing wells can be selected. These wells are cultured by limiting dilution to select for single cell clones. Single clone populations are then grown to carry out the required production campaign (Fischer et al. 2017, Barron et al. 2011A).
The miRNA miR-557 has been demonstrated by Fischer et al. to confer enhanced CHO cell growth, viability and productivity. The miR-557 is not natively expressed in CHO but is expressed in humans and was transfected into CHO. A variety of different CHO cell lines were transfected to co-express both miR-557 and different types of antibodies. These antibodies had different degrees of expression with easy to express antibodies having high expression levels and difficult to express (DTE) antibodies having low expression levels. The cells were also subject to different selection systems which ensured only cells transfected with miR-557 could grow (see Table 2).
Table 2: CHO cell lines, their selection systems, protein expressed and ease of antibody expression (adapted from Fischer et al. 2017)
| Cell line ID | Selection system | Type of protein | mAb expression level |
| CHO-mAb1 | GS | IgG antibody | High |
| CHO-mAb2 | GS | Bispecific antibody | Medium |
| CHO-mAb3 | DHFR | IgG antibody | High |
| CHO-mAb4 | GS | Bispecific IgG-scFv fusion | Medium |
| CHO-mAb5 | GS | IgG antibody | Very low |
| CHO-mAb6 | GS | IgG antibody | Low |
| CHO-mAb7 | GS | Bispecific antibody | Low |
All miR-557 transfected cell lines had universally higher viability than a negative control miRNA (miR-NT) after transient and stable expression of the miR-557. Titer was also increased compared to miR-NT. Single cell cloning was conducted for two production campaigns of CHO cells. One campaign had cells producing easy to express antibody and the other had DTE antibody. These clones were examined across different vessels ranging from 384 well plates to 6 well plates to shake flasks (the best expressing clones were selected for each up-scaling step).
Perhaps the best indicator of miR-557 applicability to industrial cell culture is where the 3 best expressing clones from each campaign were cultured in ambr™15 microbioreactor systems for 10 days. While the miR-NT cells showed roughly similar titer to the miR-557 cells for easy to express antibody (see Fig 6), there was a significant increase in titer for the DTE antibody when miR-557 was expressed (see Fig 7).
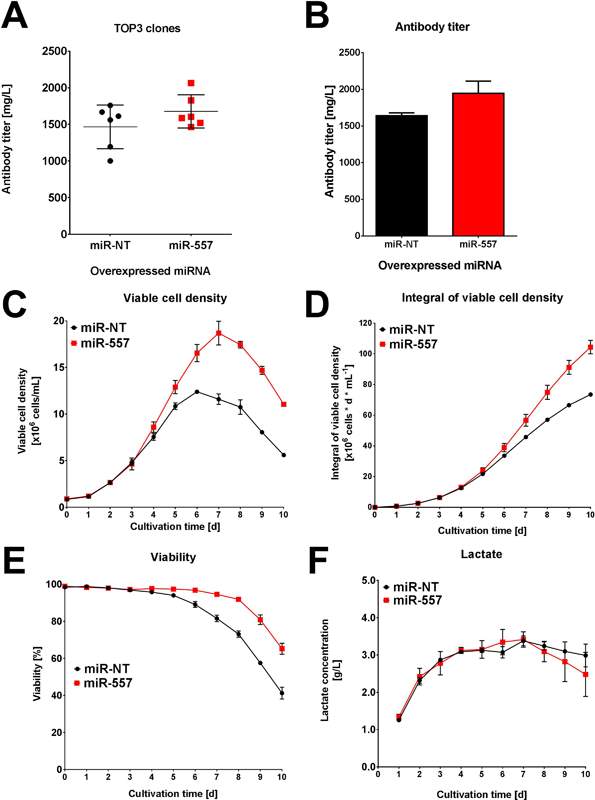
Fig 6: Performance for easy to express antibody. (A) Average titer of top 3 clones in each campaign. (B) Titer of best clone in each campaign. (C) Viable cell density of best clone in each campaign. (D) Integral viable cell density of best clone in each campaign. (E) Viability of best clone in each campaign. (F) Lactate concentration for each of the best clones in both campaigns. (Adapted from Fischer et al. 2017)
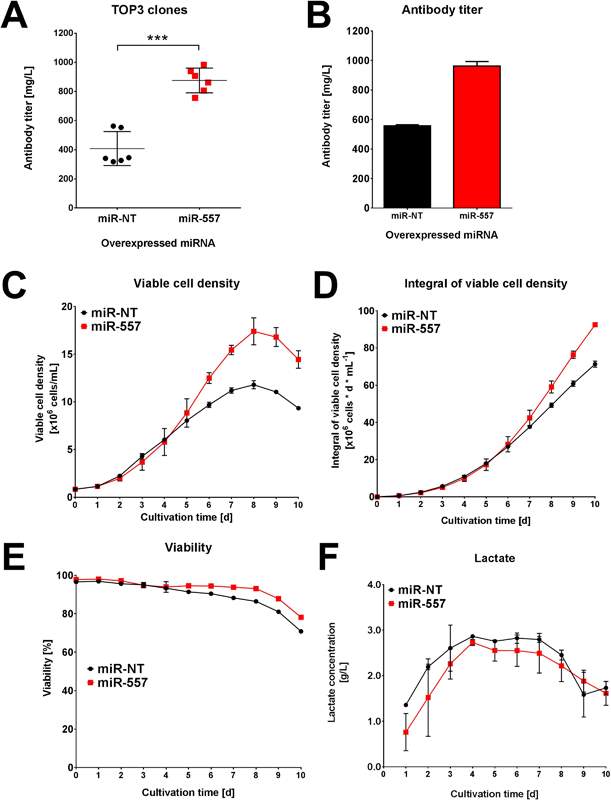
Fig 7: Performance for DTE antibody. (A) Average titer of top 3 clones in each campaign. (B) Titer of best clone in each campaign. (C) Viable cell density of best clone in each campaign. (D) Integral viable cell density of best clone in each campaign. (E) Viability of best clone in each campaign. (F) Lactate concentration for each of the best clones in both campaigns. (Adapted from Fischer et al. 2017)
As can be seen in Fig 7, the titer, viable cell density and viability from the miR-557 expressing CHO cells were markedly increased compared to miR-NT in the scaled-down industrial cell line cultivation. Product quality was later confirmed to be unaffected by miR-557 expression (Fischer et al. 2017). This is strong evidence that miRNA can be used to enhance cell culture. The miR-557 is not the only miRNA to be confirmed as beneficial to CHO cell line development. An earlier study looked at 139 miRNAs with 21% giving some benefit to CHO cell productivity. The miR-30 family in particular enhanced both productivity and cell density achieved (Fischer et al. 2014).
5. MicroRNA Sponge Biogenesis
5.1. Linear miRNA Sponge
Linear miRNA sponge transcripts are transcribed and processed in the same way as other mRNA transcripts but they can be generated in a variety of forms. The first of these are protein coding mRNAs which, when overexpressed, compete with other miRNA targets for binding. They contain the similar seed sequences in their 3’ UTR as other mRNA which are silenced by miRNA. This has been evidenced in the case of Phosphatase and Tensin Homolog (PTEN) which is a known tumour suppressor. Increased expression of the 3’ UTR of other protein coding mRNA (such as SENRINC1) results in increased expression of PTEN. The SENRINCI mRNA transcript acts a decoy and “soaks-up” miRNA which target PTEN. This prevents these miRNA from silencing PTEN expression (Tay et al. 2011).
Another form of miRNA sponge exists in the form of pseudo-genes. These are genes which, due to a mutation or deletion, no longer code for a protein. However, these genes may still be transcribed. Due to their high similarity and relatedness to other protein coding mRNA transcripts they contain target sites for miRNA binding. The pseudo-gene KRAS1 has been shown to compete with its protein coding relative, KRAS for miRNA binding (Poliseno et al. 2010).
Non-coding genes also have arguably the most significant role in generating miRNA sponge. Whilst they do not code for a protein, their transcription can significantly alter the expression of an unrelated protein coding gene. This is the case for the protein coding gene, PHO2 which reduces phosphatase levels in plants and is inhibited by mi-R399. High levels of the non-coding gene IPS1 compete with PHO2 for mi-R399 binding. IPS1 appears to be well suited to this purpose as the site where it binds to mi-R399 contains a bulge which makes it more difficult to cleave by RISC. This increases its lifespan as a potential target (Franco-Zorrilla et al. 2007).
5.2 Circular miRNA Sponge Biogenesis
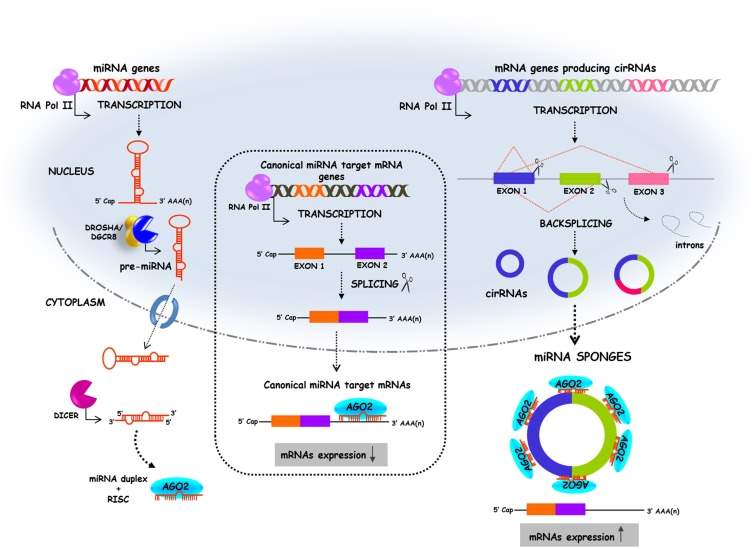
Fig 8: (left) the biogenesis pathway of linear miRNA. (centre) Canonical splicing of mRNA which is then silenced by miRNA binding. (right) “backsplicing” forming circular miRNA sponge sequences which have a number of seed sequences that miRNA can bind too. This competes with the mRNA strand for binding miRNA and thus the mRNA strand is not silenced by miRNA. (Adapted from Kulcheski, Christoff and Margis 2016)
Circular RNA (circRNA) is classed as ncRNA and was assumed to have no function until recent times (Memczak et al. 2013). Circular miRNA sponges are formed in the same way as circRNA. It is produced during the splicing stage of pre-mRNA processing and requires the same splicing machinery as canonical splicing. However, the rules followed by the splicing elements appear to be reversed and this is called “backsplicing”. In backsplicing, an upstream 3’ splicing site is joined with a downstream 5’ splicing site (Wang and Wang 2015). CircRNAs are produced co-transcriptionally and the intronic sequences which flank the sequences are believed to be key in causing circularisation. Factors which bind to these intronic sequences have been shown to modulate circRNA generation. An example of this would be the factor “muscleblind” which affected the expression of three circRNAs (Ashwal-Fluss et al. 2014). The intronic sequences contain inverted repeats which facilitate circularisation (Liang and Wilusz 2014). An example of intronic sequences include ALU repeat sequences. Flanking ALU repeats are complementary to each other and have been found in the flanking intronic sequences of many circRNA. These repeats are six times more likely to be present flanking circRNA exons compared to non-circularised RNA exons. They have been shown to contribute to circularisation which allows backsplicing to occur (see Fig 9) (Jeck et al. 2013).
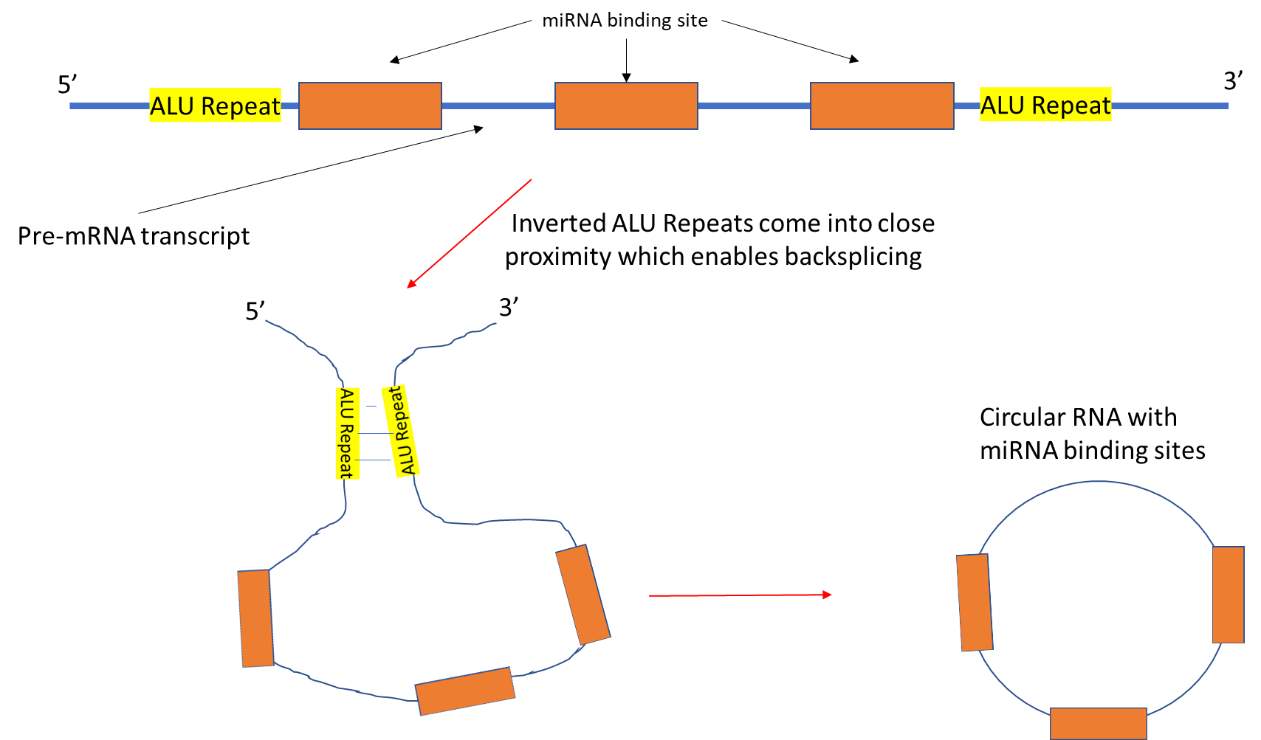
Fig 9: Pre-mRNA transcript contains sequences which are complementary to the seed sequences of miRNA. These sequences are flanked by inverted ALU repeat sequences. The ALU repeats bind to each other which brings splice sites present on exons in the pre-mRNA into proximity. This in turn allows backsplicing to occur which in forms the circRNA sponge.
The circRNA sponges are predominantly cytoplasmic and have multiple seed sequences for miRNA binding. It was reported the circRNA sponge for the microRNA, miR-7, had over 60 seed sequences which miR-7 could bind too. In this way, these circular sponges compete with miRNA targets for binding (Hansen, Kjems and Damgaard 2013, Jeck et al. 2013). Many circRNA have a seed sequence deletion at position 9-12 which introduce “wobble” and hinder cleavage fn the circRNA sponge by RISC. This prolongs the lifespan of the sponge in the cell; increasing its competitiveness (Kluiver et al. 2012).
6. Natural Functions and Applications of miRNA Sponge
6.1. Roles in disease
As much as miRNA regulate cell function and inhibit disease, there is also a natural requirement for they themselves to be regulated. As detailed earlier, this is where the various classes of miRNA sponge act in the cell. However, uncontrolled expression of a sponge can lead to disease states. Such is the case with the oncogene, PVT1, in gastric cancer. PVT1 expression has been associated with cell proliferation and is a lncRNA with 3 binding sites for the miRNA, miR-152 (Li, Meng and Yang 2017). This miRNA has previously been shown to inhibit fibroblast growth factor 2 (FGF2) which is involved in the growth and invasion of lung cancer (Cheng et al. 2014). The miR-152 also downregulates CD151 expression which aids in gastric cancer cell motility and has been shown to promote signal transduction of transforming growth factor β and hepatic growth factor in breast cancer (Zhai et al. 2014). It is therefore fair to postulate that decreased levels of miR-152 could have a role in gastric cancer proliferation. This was confirmed when Li, Meng and Yang (2017) showed that miR-152 was decreased in Gastric Cancer cells. PVT1, however, had elevated expression. This suggests that aberrant expression of PVT1, a competing endogenous RNA of targets for miR-152, can be causative of gastric cancer.
6.2 Possible Therapeutics
Since the discovery of miRNA sponges, attempts have been made to use them as potential therapies. Synthetic miRNA sponges have been created for multiple miRNA which underline disease. Some sponges even attract multiple types of miRNA. One sponge sequence has been designed in vitro to inhibit 4 miRNAs. To do this, a sequence was designed with seed MBS for binding to miR-21 (over expressed in heart failure conditions), miR-155 (known to cause immune cell differentiation) and miR-221 and miR-222 (which are paralogs with the same seed sequence and are over expressed in prostate, lung and pancreatic cancer). The sponge sequence had SanDI sites on either side (Jung et al. 2015) of the MBS. Spacers were put between the seed sequences to limit non-specific binding. This was then introduced into a plasmid with a Luciferase reporter gene where it incorporated into the 3’ UTR of the gene. The plasmid also had a SanDI restriction enzyme site previously added to allow directional cloning. This is illustrated in Fig 10.
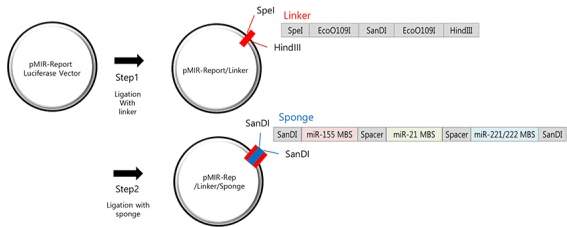
Fig 10: The linker containing the SanDI is first ligated with a reporter plasmid that expressed luciferase. Using restriction enzymes that cut at the SanDI site, the sponge can then be incorporated into the plasmid in such a way that it can be directionally cloned. (Adapted from Jung et al. 2015)
Two versions of this plasmid were introduced to 5 different cell lines which included a breast cancer cell line. One version had MBS with perfect base pair complementarity. The second had bulges or wobbles in the multiple binding sites. In all cell lines, luciferase activity was decreased as miRNA bound to the miRNA seed sequences and inhibited translation of luciferase. It was noted that only a single copy of the MBS sponge was necessary to completely inhibit luciferase for the perfect complementarity MBS sponge. However, the MBS sponge with bulged sequences required additional copy numbers to completely inhibit luciferase. This is most likely due to the wobble hindering RISC loading and cleavage; therefore inhibiting gene silencing (Kluiver et al. 2012).
The MBS sponge sequence was then added to a Red Fluorescent Protein (RFP) vector with a tetracycline inducible reporter. Again, one plasmid was used with perfectly complementary MBS and the another had bulged MBS. This was to measure whether the target miRNA were decreased after inducing expression of the sponge. It also looked at whether perfect complementarity or bulged seed sequences were superior inhibitors. It was found that the miRNAs were significantly reduced in all cell lines after inducing for sponge expression. Also, the bulged MBS sponge had a greater inhibitory effect than the perfectly complementary MBS. This was further evidenced by the increased in expression of 9 target genes which are inhibited by these miRNA. These included Stat5 for miR-221/222, Bcl-2 for miR-21 and Smad4 for miR-155. It was also noted that the sponge had strong target specificity with very little non-target miRNA binding (Jung et al. 2015). This demonstrates the possible therapeutic capabilities of miRNA sponges. Not only can they be designed to effectively target overexpressed miRNAs in diseased cells but also multiple miRNAs simultaneously.
6.3 Role in CHO Cell Engineering
Sanchez et al. (2014) have demonstrated that miRNA sponge can be used to be improve CHO cell density. Previously, Barron et al. (2011A) reported that miR-7 hindered CHO cell growth. It has since been indicated that CHO clones with high growth rates are more desirable than clones which have high specific productivity (Clarke et al. 2011). This prompted an investigation into the effects of lowering miR-7 concentrations in CHO cells and how this would affect their performance. Sanchez et al. (2014) designed a sponge sequence containing 4 bulged binding sites for miR-7. This was placed into a destabilised GFP plasmid and inserted into SEAP expressing CHO cells. Stable cell lines were established using hygromycin for 2 weeks. Fluorescence activated cell sorting (FACS) identified stable GFP expressing cells which had the miR-7 sponge integrated.
The sponge not only caused miR-7 levels to fall in the cells, but it also caused an increase in viable cell density versus a control sponge which did not have miR-7 binding sites. The sponge’s specificity for miR-7 was confirmed when additional treatment of miR-7 mimics further reduced GFP expression. CDC7 mRNA concentration, a natural target for miR-7, had a 2.5 fold increase in concentration. This is further proof that miR-7 was being re-directed from its natural targets to the sponge.
To test whether this improved CHO cell density could be mirrored in conditions closer to industrial culture, a fed batch campaign was set up for best performing miR-7 transfected clone and the best performing control clone. This was done using vented shake flasks and the campaign was run for 13 days. Viable cell density in the miR-7 sponge cells were significantly increased compared to the control sponge cells. However, the viability was not compromised and overall productivity was enhanced in miR-7 sponge cells (see Fig 11) (Sanchez et al. 2014).
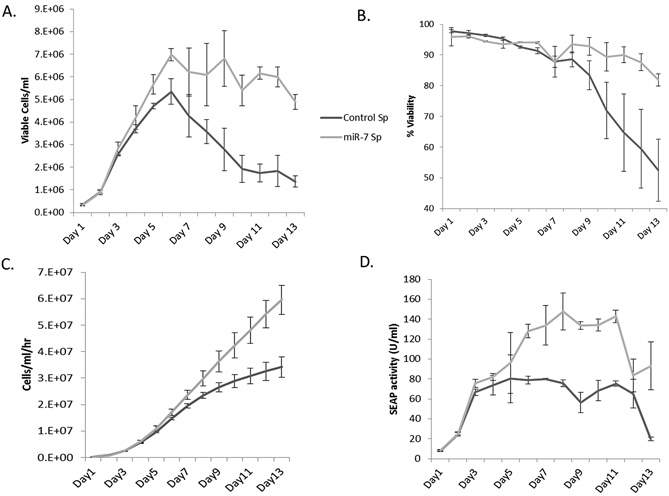
Fig 11: (A) miR-7 sponge cells achieved a higher cell density than control cells. (B)Viability was similar for the early stage of the campaign but was improved in the miR-7 sponge cells. (C) Integrated viability was calculated and it was also higher in miR-7 sponge cells. (D) Due to the increased cell density and stable viability, overall protein production was increased versus control cells. (Adapted from Sanchez et al. 2014)
This demonstrates miRNA sponge to be a potent tool in enhancing CHO cell performance. Given that culture conditions such as temperature shifts alter the productivity of CHO cells greatly (Oguchi et al. 2006), identification and manipulation of genes affected by these changes may be an avenue to maximise protein production. This is where miRNA and miRNA sponges have a significant role to play. While gene deletion using gene editing methods such as CRISPR/ Cas9 has yielded promising results in augmenting CHO Cell culture (Lee et al. 2016), the fact miRNA sponge can be induced (Jung et al. 2015) means they can be used to silence gene expression at specific points in a production campaign. This ideal as gene expression or inhibition may be desired at a specific point in a cell culture manufacturing process.
7. Conclusion
The roles of miRNA and miRNA sponge within the cell, despite being in their infancy of understanding, are becoming more and more central to gene regulation techniques. They are proof that ncRNA have a major importance within the cell. This is best highlighted by their presence in disease and how their low expression or overexpression can be critical in causing a disease. There is much to be learned from how these posttranscriptional regulators function. Indeed, being able to identify their presence and understand how they are influencing other genes in the cell can be key in identifying, distinguish and characterising a disease.
Perhaps even more exciting are the possibilities miRNA and miRNA Sponge bring to the bioprocessing world in developing host cell lines. Their ability to confer increased cell growth and productivity will be key the production of complex therapeutics. Not only does this increase the scope of what complex biologics can be manufactured, but it will also decrease the production cost of these drugs. This is often a critical hindrance in the development of drugs for the patient as companies will not manufacture a drug which isn’t profitable. If one goes beyond the world of biopharmaceuticals, it is not unfair to postulate that miRNA and miRNA sponge may even aid in the mass production of other biological products such as commercial enzymes or biofuel.
This review has looked at miRNA and miRNA sponges as classes of ncRNA. It has also examined what is known about their biogenesis and what roles they naturally have within the cell. In terms of applications, they range from biomarkers to therapeutics to cell engineering tools. Undoubtedly, this is only a fraction of their immense potential.
Experimental Design
Background
In the last 20 years, the biopharmaceutical industry has grown exponentially with much of the focus in modern drug development going towards protein based therapeutics. The Chinese Hamster Ovary (CHO) cell has established itself as the cellular workhorse for making protein based drugs due to their favourable glycosylation of proteins and its ability to be grown in suspension culture. This is in addition to their relative ease of achieving stable transgene expression (Croset et al. 2012).
However, a great deal of research has been invested into improving the productivity of CHO cells. It has been recognised that cell lines which are capable of growth to large cell densities without incurring major impact on viability are favourable cell lines for overall productivity (Clarke et al. 2011). Genetically modifying CHO cells so that they can reach these densities, without compromising their specific productivity, has become an area of focus for industry. This is where microRNA (miRNA) sponge decoys are powerful tools in CHO cell engineering.
The miRNA sponge act to inhibit miRNA which are non-coding RNA. In combination with RISC, miRNA silence gene expression through binding of complementary regions in the 3’ UTR of mRNA transcripts; blocking translation or initiating transcript degradation (Chi, Hannon and Darnell 2012). Depending on which genes they silence, miRNA can be either beneficial or inhibitory to cell growth and specific productivity. The miRNA, miR-7, is an example of a growth inhibiting miRNA. Silencing of this miRNA through sponge decoys has previously resulted in increased cell culture densities (Sanchez et al. 2014).
Sponge may exist as a linear RNA sequence or as a circular RNA (circRNA) sequence (Hansen et al. 2013). As circRNA have been found to be more stable than their linear counterparts (Li et al. 2015), it has been proposed that circRNA sponges may be superior posttranscriptional regulators. To test this hypothesis, we will examine the effect of both a naturally occurring circRNA sponge, ciRS-7, and a synthetic circRNA sponge on CHO cell growth, viability and productivity. Both of the sponges will be compared to a linear miRNA sponge in two campaigns, a batch and fed-batch campaign.
Aims/Objectives
- Determine the effect on CHO cell growth, viability and productivity of ciRS-7 versus a linear sponge.
- Quantify miR-7 and sponge expression levels within the cell
- Compare a synthetic circRNA sponge’s performance to ciRS-7
Experimental Rationale
CircRNA has been shown to have greater stability with less nuclease degradation than linear RNA. It is therefore postulated that circRNA sponge has a longer duration of effect than linear miRNA sponge. To examine if this is the case, CHO cells transfected with a linear miRNA sponge will be compared versus cells with natural ciRS-7 transfected and cells with synthetic circRNA sponge transfected. The fed-batch campaign allows industrial culture conditions to be mimicked.
Methodology
The cell line used is the CHO DP12 and it expresses a monoclonal antibody which can inhibit interleukin 8 binding (IL-8) to human neutrophil (Heinrich et al. 2011).
- CHO DP-12 cultured in 50ml TPP spin tube reactors to desired cell density with passaging every 3-4 days based on requirements.
- Naturally occurring ciRS-7 gene sequence pre-isolated and PCR amplified.
- Linear RNA sponge sequence pre-designed with multiple binding sites for miR-7a with spacers in between each binding site.
- Linear sponge sequence also has SanDI restriction enzyme sites added.
- Synthetic circRNA designed incorporating same sponge sequence as linear sponge along with inverted repeats to allow circularisation.
- Plasmid vector pcDNA3.1(+)HYG prepared by digesting with SanDI.
- Plasmid then ligated with each of the three sponge decoys in separate reactions.
- Plasmid stock is then scaled with a midi-prep to ensure there is enough DNA for transfection.
- Each type of ligated plasmid is then transfected to a CHO DP-12 culture using MIRUS-TransIT.
- Cells grown in media containing hygromycin. Cells which have integrated the plasmid into their genome will be hygromycin resistant and therefore only these cells will grow in the media.
- There will be two campaigns: a batch campaign and a fed batch campaign.
- Both campaigns will have viable cell density and cell viability measured every 24 hours by Flow Cytometry, using the Guava Easycyte programme.
- During each campaign, Trizol will be used to extract total RNA from cells.
- RNA is then purified from residual DNA.
- Total RNA then reversed transcribed to cDNA.
- RT-qPCR will quantify sponge and miR-7a gene expression to determine effectiveness of the sponge in silencing miR-7a.
- To examine overall productivity of each cell line in both campaigns, IgG ELISA will be carried out to measure monoclonal anti-IL-8.
Contingencies/Alternative Strategies
- Should the synthetic sponge sequence used have low specificity for miR-7, a different sequence or alterations to the current sequence can be implemented.
- The miRNA sponge expression level may be low due to where the sponge transgene has integrated in the cell’s genome i.e. it has integrated in a region that is not transcriptionally active. A different selection marker such as neomycin or zeocin can be used to screen for cells where this has happened. This selects cells where a favourable integration has occurred and may be a stricter screening process than Hygromycin.
- Should the sponge appear to have little to no reduction on the cell’s miR-7 levels, larger vectors that have additional multiple binding sites for miR-7 can be used.
- If sponge expression is poor or there is little inhibition of miR-7, we will see if this is recurring in other CHO cell lines.
- Sponge expression or inhibition of miR-7 may also be affected by the media the cells are cultured in. In this instance, alternative culture medias will be explored.
- If there is poor transfection efficiency, a different transfection method may be used e.g. lipofectamine.
Bibliography
- Ashwal-Fluss, R., Meyer, M., Pamudurti, N.R., Ivanov, A., Bartok, O., Hanan, M., Evantal, N., Memczak, S., Rajewsky, N. and Kadener, S. 2014. CircRNA Biogenesis Competes with Pre-mRNA Splicing. Molecular Cell, 56(1), pp.55–66.
- Barron, N., Kumar, N., Sanchez, N., Doolan, P., Clarke, C., Meleady, P., O’Sullivan, F. and Clynes, M. 2011. Engineering CHO cell growth and recombinant protein productivity by overexpression of miR-7. Journal of Biotechnology, 151(2), pp.204–211.
- Barron, N., Sanchez, N., Kelly, P. and Clynes, M. 2011. MicroRNAs: tiny targets for engineering CHO cell phenotypes? Biotechnology Letters, 33(1), pp.11–21.
- Batisse, J., Batisse, C., Budd, A., Böttcher, B. and Hurt, E. 2009. Purification of Nuclear Poly(A)-binding Protein Nab2 Reveals Association with the Yeast Transcriptome and a Messenger Ribonucleoprotein Core Structure. The Journal of Biological Chemistry, 284(50), pp.34911–34917.
- Blahna, M.T. and Hata, A. 2012. Smad-mediated regulation of microRNA biosynthesis. FEBS Letters, 586(14), pp.1906–1912.
- Brennecke, J., Aravin, A.A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R. and Hannon, G.J. 2007. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell, 128(6), pp.1089–1103.
- Cai, X., Hagedorn, C.H. and Cullen, B.R. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA, 10(12), pp.1957–1966.
- Carthew, R.W. and Sontheimer, E.J. 2009. Origins and Mechanisms of miRNAs and siRNAs. Cell, 136(4), pp.642–655.
- Chakraborty, C., Sharma, A.R., Sharma, G., Doss, C.G.P. and Lee, S.-S. 2017. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Molecular Therapy. Nucleic Acids, 8, pp.132–143.
- Cheloufi, S., Dos Santos, C.O., Chong, M.M.W. and Hannon, G.J. 2010. A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature, 465(7298), pp.584–589.
- Chen, C.-Y.A. and Shyu, A.-B. 2011. Mechanisms of deadenylation-dependent decay. Wiley interdisciplinary reviews. RNA, 2(2), pp.167–183.
- Chen, Z., Wu, J., Yang, C., Fan, P., Balazs, L., Jiao, Y., Lu, M., Gu, W., Li, C., Pfeffer, L.M., Tigyi, G. and Yue, J. 2012. DiGeorge Syndrome Critical Region 8 (DGCR8) Protein-mediated microRNA Biogenesis Is Essential for Vascular Smooth Muscle Cell Development in Mice. The Journal of Biological Chemistry, 287(23), pp.19018–19028.
- Chendrimada, T.P., Gregory, R.I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K. and Shiekhattar, R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature, 436(7051), pp.740–744.
- Cheng, Z., Ma, R., Tan, W. and Zhang, L. 2014. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Experimental & Molecular Medicine, 46(9), p.e112.
- Chi, S.W., Hannon, G.J. and Darnell, R.B. 2012. An alternative mode of microRNA target recognition. Nature Structural and Molecular Biology, 19(3), pp.321–327.
- Chorn, G., Klein-McDowell, M., Zhao, L., Saunders, M.A., Flanagan, W.M., Willingham, A.T. and Lim, L.P. 2012. Single-stranded microRNA mimics. RNA, 18(10), pp.1796–1804.
- Clarke, C., Doolan, P., Barron, N., Meleady, P., O’Sullivan, F., Gammell, P., Melville, M., Leonard, M. and Clynes, M. 2011. Large scale microarray profiling and coexpression network analysis of CHO cells identifies transcriptional modules associated with growth and productivity. Journal of Biotechnology, 155(3), pp.350–359.
- Croset, A., Delafosse, L., Gaudry, J.-P., Arod, C., Glez, L., Losberger, C., Begue, D., Krstanovic, A., Robert, F., Vilbois, F., Chevalet, L. and Antonsson, B. 2012. Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. Journal of Biotechnology, 161(3), pp.336–348.
- Daffis, S., Szretter, K.J., Schriewer, J., Li, J., Youn, S., Errett, J., Lin, T.-Y., Schneller, S., Zust, R., Dong, H., Thiel, V., Pierson, T.C., Buller, R.M., Gale, M., Shi, P.-Y. and Diamond, M.S. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature, 468(7322), pp.452–456.
- Davis, B.N., Hilyard, A.C., Lagna, G. and Hata, A. 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature, 454(7200), pp.56–61.
- Davis, B.N., Hilyard, A.C., Nguyen, P.H., Lagna, G. and Hata, A. 2010. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Molecular cell, 39(3), pp.373–384.
- Fabian, M.R., Cieplak, M.K., Frank, F., Morita, M., Green, J., Srikumar, T., Nagar, B., Yamamoto, T., Raught, B., Duchaine, T.F. and Sonenberg, N. 2011. MiRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nature Structural & Molecular Biology, 18(11), pp.1211–1217.
- Fischer, S., Buck, T., Wagner, A., Ehrhart, C., Giancaterino, J., Mang, S., Schad, M., Mathias, S., Aschrafi, A., Handrick, R. and Otte, K. 2014. A functional high-content miRNA screen identifies miR-30 family to boost recombinant protein production in CHO cells. Biotechnology Journal, 9(10), pp.1279–1292.
- Fischer, S., Marquart, K.F., Pieper, L.A., Fieder, J., Gamer, M., Gorr, I., Schulz, P. and Bradl, H. 2017. MiRNA engineering of CHO cells facilitates production of difficult-to-express proteins and increases success in cell line development. Biotechnology and Bioengineering, 114(7), pp.1495–1510.
- Flamand, M.N., Wu, E., Vashisht, A., Jannot, G., Keiper, B.D., Simard, M.J., Wohlschlegel, J. and Duchaine, T.F. 2016. Poly(A)-binding proteins are required for microRNA-mediated silencing and to promote target deadenylation in C. elegans. Nucleic Acids Research, 44(12), pp.5924–5935.
- Franco-Zorrilla, J.M., Valli, A., Todesco, M., Mateos, I., Puga, M.I., Rubio-Somoza, I., Leyva, A., Weigel, D., García, J.A. and Paz-Ares, J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics, 39(8), p.ng2079.
- Frank, F., Sonenberg, N. and Nagar, B. 2010. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature, 465(7299), pp.818–822.
- Fukuda, T., Yamagata, K., Fujiyama, S., Matsumoto, T., Koshida, I., Yoshimura, K., Mihara, M., Naitou, M., Endoh, H., Nakamura, T., Akimoto, C., Yamamoto, Y., Katagiri, T., Foulds, C., Takezawa, S., Kitagawa, H., Takeyama, K., O’Malley, B.W. and Kato, S. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nature Cell Biology, 9(5), pp.604–611.
- Goldgraben, M.A., Russell, R., Rueda, O.M., Caldas, C. and Git, A. 2016. Double-stranded microRNA mimics can induce length- and passenger strand–dependent effects in a cell type–specific manner. RNA, 22(2), pp.193–203.
- Gregory, R.I., Kai-Ping Yan, Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N. and Shiekhattar, R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature, 432(7014), pp.235–240.
- Gu, S., Jin, L., Huang, Y., Zhang, F. and Kay, M.A. 2012. Slicing-Independent RISC Activation Requires the Argonaute PAZ Domain. Current Biology, 22(16), pp.1536–1542.
- Han, J., Lee, Y., Yeom, K.-H., Nam, J.-W., Heo, I., Rhee, J.-K., Sohn, S.Y., Cho, Y., Zhang, B.-T. and Kim, V.N. 2006. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell, 125(5), pp.887–901.
- Han, J., Pedersen, J.S., Kwon, S.C., Belair, C.D., Kim, Y.-K., Yeom, K.-H., Yang, W.-Y., Haussler, D., Blelloch, R. and Kim, V.N. 2009. Posttranscriptional Crossregulation between Drosha and DGCR8. Cell, 136(1), pp.75–84.
- Hansen, T.B., Jensen, T.I., Clausen, B.H., Bramsen, J.B., Finsen, B., Damgaard, C.K. and Kjems, J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature, 495(7441), pp.384–388.
- Hansen, T.B., Kjems, J. and Damgaard, C.K. 2013. Circular RNA and miR-7 in Cancer. Cancer Research, 73(18), pp.5609–5612.
- Heinrich, C., Wolf, T., Kropp, C., Northoff, S. and Noll, T. 2011. Growth characterization of CHO DP-12 cell lines with different high passage histories. BMC Proceedings, 5(Suppl 8), p.P29.
- Horman, S.R., Janas, M.M., Litterst, C., Wang, B., MacRae, I.J., Sever, M.J., Morrissey, D.V., Graves, P., Luo, B., Umesalma, S., Qi, H.H., Miraglia, L.J., Novina, C.D. and Orth, A.P. 2013. Akt-Mediated Phosphorylation of Argonaute 2 Downregulates Cleavage and Upregulates Translational Repression of MicroRNA Targets. Molecular Cell, 50(3), pp.356–367.
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G. and Linsley, P.S. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology, 21(6), p.nbt831.
- Jeck, W.R., Sorrentino, J.A., Wang, K., Slevin, M.K., Burd, C.E., Liu, J., Marzluff, W.F. and Sharpless, N.E. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19(2), pp.141–157.
- Jiang, L., Shao, C., Wu, Q.-J., Chen, G., Zhou, J., Yang, B., Li, H., Gou, L.-T., Zhang, Y., Wang, Y., Yeo, G.W., Zhou, Y. and Fu, X.-D. 2017. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nature Structural & Molecular Biology, 24(10), pp.816–824.
- Jin, H.Y., Gonzalez-Martin, A., Miletic, A.V., Lai, M., Knight, S., Sabouri-Ghomi, M., Head, S.R., Macauley, M.S., Rickert, R.C. and Xiao, C. 2015. Transfection of microRNA Mimics Should Be Used with Caution. Frontiers in Genetics, 6.
- Jung, J., Yeom, C., Choi, Y.-S., Kim, S., Lee, E., Park, M.J., Kang, S.W., Kim, S.B. and Chang, S. 2015. Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget, 6(24), pp.20370–20387.
- Juvvuna, P.K., Khandelia, P., Lee, L.M. and Makeyev, E.V. 2012. Argonaute identity defines the length of mature mammalian microRNAs. Nucleic Acids Research, 40(14), pp.6808–6820.
- Kim, Y.-K., Kim, B. and Kim, V.N. 2016. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proceedings of the National Academy of Sciences of the United States of America, 113(13), pp.E1881–E1889.
- Kluiver, J., Slezak-Prochazka, I., Smigielska-Czepiel, K., Halsema, N., Kroesen, B.-J. and van den Berg, A. 2012. Generation of miRNA sponge constructs. Methods, 58(2), pp.113–117.
- Krützfeldt, J., Rajewsky, N., Braich, R., Rajeev, K.G., Tuschl, T., Manoharan, M. and Stoffel, M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature, 438(7068), pp.685–689.
- Kulcheski, F.R., Christoff, A.P. and Margis, R. 2016. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. Journal of Biotechnology, 238(Supplement C), pp.42–51.
- Kwak, P.B. and Tomari, Y. 2012. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nature Structural & Molecular Biology, 19(2), pp.145–151.
- Lam, J.K.W., Chow, M.Y.T., Zhang, Y. and Leung, S.W.S. 2015. SiRNA Versus miRNA as Therapeutics for Gene Silencing. Molecular Therapy. Nucleic Acids, 4(9), p.e252.
- Latreille, M., Herrmanns, K., Renwick, N., Tuschl, T., Malecki, M.T., McCarthy, M.I., Owen, K.R., Rülicke, T. and Stoffel, M. 2015. MiR-375 gene dosage in pancreatic β-cells: implications for regulation of β-cell mass and biomarker development. Journal of Molecular Medicine, 93(10), pp.1159–1169.
- Lau, P.-W., Guiley, K.Z., De, N., Potter, C.S., Carragher, B. and MacRae, I.J. 2012. The Molecular Architecture of Human Dicer. Nature structural & molecular biology, 19(4), pp.436–440.
- Le Thomas, A., Toth, K. and Aravin, A. 2014. To be or not to be a piRNA: Genomic origin and processing of piRNAs. Genome biology, 15, p.204.
- Lee, H.Y. and Doudna, J.A. 2012. TRBP alters human precursor microRNA processing in vitro. RNA, 18(11), pp.2012–2019.
- Lee, H.Y., Zhou, K., Smith, A.M., Noland, C.L. and Doudna, J.A. 2013. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Research, 41(13), pp.6568–6576.
- Lee, N., Shin, J., Park, J.H., Lee, G.M., Cho, S. and Cho, B.-K. 2016. Targeted Gene Deletion Using DNA-Free RNA-Guided Cas9 Nuclease Accelerates Adaptation of CHO Cells to Suspension Culture. ACS Synthetic Biology, 5(11), pp.1211–1219.
- Lee, R.C., Feinbaum, R.L. and Ambros, V. 1993. The C. elegans heterochronic gene lin- 4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75(5), pp.843–854.
- Lee, S.J., Jiko, C., Yamashita, E. and Tsukihara, T. 2011. Selective nuclear export mechanism of small RNAs. Current Opinion in Structural Biology, 21(1), pp.101–108.
- Lennox, K.A., Owczarzy, R., Thomas, D.M., Walder, J.A. and Behlke, M.A. 2013. Improved Performance of Anti-miRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Molecular Therapy – Nucleic Acids, 2(Supplement C), p.e117.
- Li, T., Meng, X. and Yang, W. 2017. Long Noncoding RNA PVT1 Acts as a “Sponge” to Inhibit microRNA-152 in Gastric Cancer Cells. Digestive Diseases and Sciences, 62(11), pp.3021–3028.
- Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., Chen, D., Gu, J., He, X. and Huang, S. 2015. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Research, 25(8), p.981.
- Liang, D. and Wilusz, J.E. 2014. Short intronic repeat sequences facilitate circular RNA production. Genes & Development, 28(20), pp.2233–2247.
- Lim, M.Y.T., Ng, A.W.T., Chou, Y., Lim, T.P., Simcox, A., Tucker-Kellogg, G. and Okamura, K. 2016. The Drosophila Dicer-1 Partner Loquacious Enhances miRNA Processing from Hairpins with Unstable Structures at the Dicing Site. Cell Reports, 15(8), pp.1795–1808.
- Liu, Q., Gao, Q., Zhang, Y., Li, Z. and Mei, X. 2017. MicroRNA-590 promotes pathogenic Th17 cell differentiation through targeting Tob1 and is associated with multiple sclerosis. Biochemical and Biophysical Research Communications, 493(2), pp.901–908.
- Luo, J., Nikolaev, A.Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L. and Gu, W. 2001. Negative Control of p53 by Sir2α Promotes Cell Survival under Stress. Cell, 107(2), pp.137–148.
- Lytle, J.R., Yario, T.A. and Steitz, J.A. 2007. Target mRNAs Are Repressed as Efficiently by microRNA-Binding Sites in the 5’ UTR as in the 3’ UTR. Proceedings of the National Academy of Sciences of the United States of America, 104(23), pp.9667–9672.
- Ma, J.-B., Ye, K. and Patel, D.J. 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature, 429(6989), p.nature02519.
- MacRae, I.J., Zhou, K. and Doudna, J.A. 2007. Structural determinants of RNA recognition and cleavage by Dicer. Nature Structural and Molecular Biology, 14(10), p.nsmb1293.
- Martinez, N.J. and Gregory, R.I. 2013. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA, 19(5), pp.605–612.
- Mathis, D., Vence, L. and Benoist, C. 2001. Beta-Cell death during progression to diabetes. Nature, 414(6865), p.792.
- Matsui, M., Prakash, T.P. and Corey, D.R. 2016. Argonaute 2-dependent Regulation of Gene Expression by Single-stranded miRNA Mimics. Molecular Therapy, 24(5), pp.946–955.
- Maurer, P., Redd, M., Solsbacher, J., Bischoff, F.R., Greiner, M., Podtelejnikov, A.V., Mann, M., Stade, K., Weis, K. and Schlenstedt, G. 2001. The Nuclear Export Receptor Xpo1p Forms Distinct Complexes with NES Transport Substrates and the Yeast Ran Binding Protein 1 (Yrb1p). Molecular Biology of the Cell, 12(3), pp.539–549.
- Meister, G. 2013. Argonaute proteins: functional insights and emerging roles. Nature Reviews Genetics, 14(7), p.nrg3462.
- Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., Maier, L., Mackowiak, S.D., Gregersen, L.H., Munschauer, M., Loewer, A., Ziebold, U., Landthaler, M., Kocks, C., Noble, F. le and Rajewsky, N. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), p.333.
- Nolan, C.J., Damm, P. and Prentki, M. 2011. Type 2 diabetes across generations: from pathophysiology to prevention and management. The Lancet, 378(9786), pp.169–181.
- Noland, C.L., Ma, E. and Doudna, J.A. 2011. SiRNA Repositioning for Guide Strand Selection by Human Dicer Complexes. Molecular Cell, 43(1), pp.110–121.
- Oguchi, S., Saito, H., Tsukahara, M. and Tsumura, H. 2006. PH Condition in temperature shift cultivation enhances cell longevity and specific hMab productivity in CHO culture. Cytotechnology, 52(3), pp.199–207.
- Okada, C., Yamashita, E., Lee, S.J., Shibata, S., Katahira, J., Nakagawa, A., Yoneda, Y. and Tsukihara, T. 2009. A High-Resolution Structure of the Pre-microRNA Nuclear Export Machinery. Science, 326(5957), pp.1275–1279.
- Park, J.-E., Heo, I., Tian, Y., Simanshu, D.K., Chang, H., Jee, D., Patel, D.J. and Kim, V.N. 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature, 475(7355), pp.201–205.
- Park, J.H. and Shin, C. 2015a. Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Research, 43(19), pp.9418–9433.
- Park, J.H. and Shin, C. 2015b. Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Research, 43(19), pp.9418–9433.
- Pasquinelli, A.E., Reinhart, B.J., Slack, F., Martindale, M.Q., Kuroda, M.I., Maller, B., Hayward, D.C., Ball, E.E., Degnan, B., Muller, P., Spring, J., Srinivasan, A., Fishman, M., Finnerty, J., Corbo, J., Levine, M., Leahy, P., Davidson, E. and Ruvkun, G. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 408(6808), p.86.
- Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W.J. and Pandolfi, P.P. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature, 465(7301), pp.1033–1038.
- Poy, M.N., Hausser, J., Trajkovski, M., Braun, M., Collins, S., Rorsman, P., Zavolan, M., Stoffel, M. and Lodish, H.F. 2009. MiR-375 Maintains Normal Pancreatic α- and β-Cell Mass. Proceedings of the National Academy of Sciences of the United States of America, 106(14), pp.5813–5818.
- Pradère, U., Halloy, F. and Hall, J. 2017. Chemical synthesis of long RNAs with terminal 5′-phosphate groups. Chemistry – A European Journal, 23(22), pp.5210–5213.
- Rupaimoole, R. and Slack, F.J. 2017. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews Drug Discovery, 16(3), p.203.
- Saito, K., Ishizuka, A., Siomi, H. and Siomi, M.C. 2005. Processing of Pre-microRNAs by the Dicer-1–Loquacious Complex in Drosophila Cells. PLoS Biology, 3(7).
- Samandari, N., Mirza, A.H., Nielsen, L.B., Kaur, S., Hougaard, P., Fredheim, S., Mortensen, H.B. and Pociot, F. 2017. Circulating microRNA levels predict residual beta cell function and glycaemic control in children with type 1 diabetes mellitus. Diabetologia, 60(2), pp.354–363.
- Sanchez, N., Kelly, P., Gallagher, C., Lao, N.T., Clarke, C., Clynes, M. and Barron, N. 2014. CHO cell culture longevity and recombinant protein yield are enhanced by depletion of miR-7 activity via sponge decoy vectors. Biotechnology Journal, 9(3), pp.396–404.
- Suzuki, H.I., Yamagata, K., Sugimoto, K., Iwamoto, T., Kato, S. and Miyazono, K. 2009. Modulation of microRNA processing by p53. Nature, 460(7254), pp.529–533.
- Tay, Y., Kats, L., Salmena, L., Weiss, D., Tan, S.M., Ala, U., Karreth, F., Poliseno, L., Provero, P., Di Cunto, F., Lieberman, J., Rigoutsos, I. and Pandolfi, P.P. 2011. Coding-Independent Regulation of the Tumor Suppressor PTEN by Competing Endogenous mRNAs. Cell, 147(2), pp.344–357.
- Wang, Y. and Wang, Z. 2015. Efficient backsplicing produces translatable circular mRNAs. RNA, 21(2), pp.172–179.
- Wu, Q., Song, R., Ortogero, N., Zheng, H., Evanoff, R., Small, C.L., Griswold, M.D., Namekawa, S.H., Royo, H., Turner, J.M. and Yan, W. 2012. The RNase III Enzyme DROSHA Is Essential for MicroRNA Production and Spermatogenesis. The Journal of Biological Chemistry, 287(30), pp.25173–25190.
- Xiol, J., Cora, E., Koglgruber, R., Chuma, S., Subramanian, S., Hosokawa, M., Reuter, M., Yang, Z., Berninger, P., Palencia, A., Benes, V., Penninger, J., Sachidanandam, R. and Pillai, R.S. 2012. A Role for Fkbp6 and the Chaperone Machinery in piRNA Amplification and Transposon Silencing. Molecular Cell, 47(6), pp.970–979.
- Yamakuchi, M., Ferlito, M. and Lowenstein, C.J. 2008. MiR-34a repression of SIRT1 regulates apoptosis. Proceedings of the National Academy of Sciences of the United States of America, 105(36), pp.13421–13426.
- Yang, Z., Chen, K.-M., Pandey, R.R., Homolka, D., Reuter, M., Janeiro, B.K.R., Sachidanandam, R., Fauvarque, M.-O., McCarthy, A.A. and Pillai, R.S. 2016. PIWI Slicing and EXD1 Drive Biogenesis of Nuclear piRNAs from Cytosolic Targets of the Mouse piRNA Pathway. Molecular Cell, 61(1), pp.138–152.
- Yoontae Lee, Chiyoung Ahn, Jinju Han, Hyounjeong Choi, Jaekwang Kim, Jeongbin Yim, Junho Lee, Provost, P., Rådmark, O., Sunyoung Kim and Kim, N. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature, 425(6956), p.415.
- Zekri, L., Kuzuoğlu-Öztürk, D. and Izaurralde, E. 2013. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. The EMBO Journal, 32(7), pp.1052–1065.
- Zhai, R., Kan, X., Wang, B., Du, H., Long, Y., Wu, H., Tao, K., Wang, G., Bao, L., Li, F. and Zhang, W. 2014. MiR-152 suppresses gastric cancer cell proliferation and motility by targeting CD151. Tumor Biology, 35(11), pp.11367–11373.
- Zhang, Z., Xu, J., Koppetsch, B.S., Wang, J., Tipping, C., Ma, S., Weng, Z., Theurkauf, W.E. and Zamore, P.D. 2011. Heterotypic piRNA Ping-Pong Requires Qin, a Protein with Both E3 ligase and Tudor Domains. Molecular cell, 44(4), pp.572–584.
- Zipprich, J.T., Bhattacharyya, S., Mathys, H. and Filipowicz, W. 2009. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA, 15(5), pp.781–793.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Biomedical Science"
Biomedical Science focuses on how cells, organs and systems function in the human body and underpins much of modern medicine. Biomedical Science applies parts of natural and/or formal sciences to help develop advances in healthcare.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




